Host restriction factors in retroviral infection: promises in virus-host interaction
- PMID: 23254112
- PMCID: PMC3549941
- DOI: 10.1186/1742-4690-9-112
Host restriction factors in retroviral infection: promises in virus-host interaction
Abstract
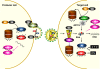
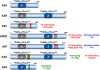
Retroviruses have an intricate life cycle. There is much to be learned from studying retrovirus-host interactions. Among retroviruses, the primate lentiviruses have one of the more complex genome structures with three categories of viral genes: structural, regulatory, and accessory genes. Over time, we have gained increasing understanding of the lentivirus life cycle from studying host factors that support virus replication. Similarly, studies on host restriction factors that inhibit viral replication have also made significant contributions to our knowledge. Here, we review recent progress on the rapidly growing field of restriction factors, focusing on the antiretroviral activities of APOBEC3G, TRIM5, tetherin, SAMHD1, MOV10, and cellular microRNAs (miRNAs), and the counter-activities of Vif, Vpu, Vpr, Vpx, and Nef.
Figures





References
-
- Lilly F. Susceptibility to two strains of friend leukemia virus in mice. Science. 1967;155(761):461–462. - PubMed
-
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382(6594):826–829. - PubMed
-
- Kozak CA, Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225(2):300–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

