Regulation of error-prone translesion synthesis by Spartan/C1orf124
- PMID: 23254330
- PMCID: PMC3561950
- DOI: 10.1093/nar/gks1267
Regulation of error-prone translesion synthesis by Spartan/C1orf124
Abstract
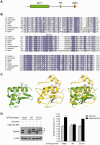
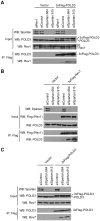
Translesion synthesis (TLS) employs low fidelity polymerases to replicate past damaged DNA in a potentially error-prone process. Regulatory mechanisms that prevent TLS-associated mutagenesis are unknown; however, our recent studies suggest that the PCNA-binding protein Spartan plays a role in suppression of damage-induced mutagenesis. Here, we show that Spartan negatively regulates error-prone TLS that is dependent on POLD3, the accessory subunit of the replicative DNA polymerase Pol δ. We demonstrate that the putative zinc metalloprotease domain SprT in Spartan directly interacts with POLD3 and contributes to suppression of damage-induced mutagenesis. Depletion of Spartan induces complex formation of POLD3 with Rev1 and the error-prone TLS polymerase Pol ζ, and elevates mutagenesis that relies on POLD3, Rev1 and Pol ζ. These results suggest that Spartan negatively regulates POLD3 function in Rev1/Pol ζ-dependent TLS, revealing a previously unrecognized regulatory step in error-prone TLS.
Figures


References
-
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. - PubMed
-
- Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. - PubMed
-
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

