Binding to the DNA minor groove by heterocyclic dications: from AT-specific monomers to GC recognition with dimers
- PMID: 23255206
- PMCID: PMC3539175
- DOI: 10.1002/0471142700.nc0808s51
Binding to the DNA minor groove by heterocyclic dications: from AT-specific monomers to GC recognition with dimers
Abstract
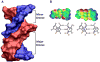
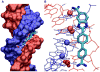
Compounds that bind in the DNA minor groove have provided critical information on DNA molecular recognition, have found extensive uses in biotechnology, and are providing clinically useful drugs against diseases as diverse as cancer and sleeping sickness. This review focuses on the development of clinically useful heterocyclic diamidine minor groove binders. These compounds have shown us that the classical model for minor groove binding in AT DNA sequences must be expanded in several ways: compounds with nonstandard shapes can bind strongly to the groove, water can be directly incorporated into the minor groove complex in an interfacial interaction, and the compounds can form cooperative stacked dimers to recognize GC and mixed AT/GC base pair sequences.
© 2012 by John Wiley & Sons, Inc.
Figures








Similar articles
-
Binding to the DNA Minor Groove by Heterocyclic Dications: from AT Specific to GC Recognition Compounds.Curr Protoc. 2023 Apr;3(4):e729. doi: 10.1002/cpz1.729. Curr Protoc. 2023. PMID: 37071034 Review.
-
Out-of-shape DNA minor groove binders: induced fit interactions of heterocyclic dications with the DNA minor groove.Biochemistry. 2005 Nov 15;44(45):14701-8. doi: 10.1021/bi051791q. Biochemistry. 2005. PMID: 16274217
-
Evaluation of the influence of compound structure on stacked-dimer formation in the DNA minor groove.Biochemistry. 2001 Feb 27;40(8):2511-21. doi: 10.1021/bi002301r. Biochemistry. 2001. PMID: 11327873
-
Alternative Approach to Sequence-Specific Recognition of DNA: Cooperative Stacking of Dication Dimers─Sensitivity to Compound Curvature, Aromatic Structure, and DNA Sequence.ACS Chem Biol. 2025 Feb 21;20(2):489-506. doi: 10.1021/acschembio.4c00800. Epub 2025 Feb 7. ACS Chem Biol. 2025. PMID: 39920086 Free PMC article.
-
A New Generation of Minor-Groove-Binding-Heterocyclic Diamidines That Recognize G·C Base Pairs in an AT Sequence Context.Molecules. 2019 Mar 7;24(5):946. doi: 10.3390/molecules24050946. Molecules. 2019. PMID: 30866557 Free PMC article. Review.
Cited by
-
Single GC base pair recognition by a heterocyclic diamidine: structures, affinities, and dynamics.RSC Adv. 2024 Sep 18;14(40):29675-29682. doi: 10.1039/d4ra05957c. eCollection 2024 Sep 12. RSC Adv. 2024. PMID: 39297050 Free PMC article.
-
A modular design for minor groove binding and recognition of mixed base pair sequences of DNA.Chem Commun (Camb). 2017 Sep 25;53(75):10406-10409. doi: 10.1039/c7cc06246j. Epub 2017 Sep 7. Chem Commun (Camb). 2017. PMID: 28880316 Free PMC article.
-
The Thiophene "Sigma-Hole" as a Concept for Preorganized, Specific Recognition of G⋅C Base Pairs in the DNA Minor Groove.Chemistry. 2016 Oct 17;22(43):15404-15412. doi: 10.1002/chem.201603422. Epub 2016 Sep 14. Chemistry. 2016. PMID: 27624927 Free PMC article.
-
Design and synthesis of heterocyclic cations for specific DNA recognition: from AT-rich to mixed-base-pair DNA sequences.J Org Chem. 2014 Feb 7;79(3):852-66. doi: 10.1021/jo402599s. Epub 2014 Jan 21. J Org Chem. 2014. PMID: 24422528 Free PMC article.
-
Imino proton NMR guides the reprogramming of A•T specific minor groove binders for mixed base pair recognition.Nucleic Acids Res. 2016 Jun 2;44(10):4519-27. doi: 10.1093/nar/gkw353. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131382 Free PMC article.
References
-
- Bailly C, Tardy C, Wang L, Armitage B, Hopkins K, Kumar A, Schuster GB, Boykin DW, Wilson WD. Recognition of ATGA sequences by the unfused aromatic dication DB293 forming stacked dimers in the DNA minor groove. Biochemistry. 2001;40:9770–9779. - PubMed
-
- Bajic M, Kumar A, Boykin DW. Synthesis of 2,5-bis-(4-cyanophenyl)-furan. Heterocycl Commun. 1996;2:135–140.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

