Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila
- PMID: 23255357
- PMCID: PMC3538939
- DOI: 10.4049/jimmunol.1102486
Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila
Abstract
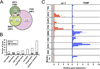
The fruit fly Drosophila melanogaster is a good model to unravel the molecular mechanisms of innate immunity and has led to some important discoveries about the sensing and signaling of microbial infections. The response of Drosophila to virus infections remains poorly characterized and appears to involve two facets. On the one hand, RNA interference involves the recognition and processing of dsRNA into small interfering RNAs by the host RNase Dicer-2 (Dcr-2), whereas, on the other hand, an inducible response controlled by the evolutionarily conserved JAK-STAT pathway contributes to the antiviral host defense. To clarify the contribution of the small interfering RNA and JAK-STAT pathways to the control of viral infections, we have compared the resistance of flies wild-type and mutant for Dcr-2 or the JAK kinase Hopscotch to infections by seven RNA or DNA viruses belonging to different families. Our results reveal a unique susceptibility of hop mutant flies to infection by Drosophila C virus and cricket paralysis virus, two members of the Dicistroviridae family, which contrasts with the susceptibility of Dcr-2 mutant flies to many viruses, including the DNA virus invertebrate iridescent virus 6. Genome-wide microarray analysis confirmed that different sets of genes were induced following infection by Drosophila C virus or by two unrelated RNA viruses, Flock House virus and Sindbis virus. Overall, our data reveal that RNA interference is an efficient antiviral mechanism, operating against a large range of viruses, including a DNA virus. By contrast, the antiviral contribution of the JAK-STAT pathway appears to be virus specific.
Figures


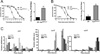
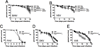
References
-
- Beutler B, Eidenschenk C, Crozat K, Imler JL, Takeuchi O, Hoffmann JA, Akira S. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. - PubMed
-
- Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, Garcia-Sastre A, Compans R, Pulendran B. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–547. - PMC - PubMed
-
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

