Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution
- PMID: 23255793
- PMCID: PMC3571365
- DOI: 10.1128/JVI.00776-12
Negevirus: a proposed new taxon of insect-specific viruses with wide geographic distribution
Abstract
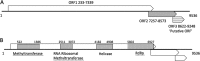
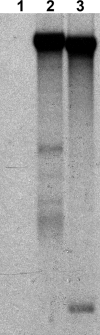
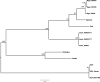
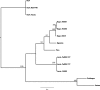
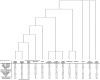
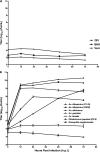
Six novel insect-specific viruses, isolated from mosquitoes and phlebotomine sand flies collected in Brazil, Peru, the United States, Ivory Coast, Israel, and Indonesia, are described. Their genomes consist of single-stranded, positive-sense RNAs with poly(A) tails. By electron microscopy, the virions appear as spherical particles with diameters of ∼45 to 55 nm. Based on their genome organization and phylogenetic relationship, the six viruses, designated Negev, Ngewotan, Piura, Loreto, Dezidougou, and Santana, appear to form a new taxon, tentatively designated Negevirus. Their closest but still distant relatives are citrus leposis virus C (CiLV-C) and viruses in the genus Cilevirus, which are mite-transmitted plant viruses. The negeviruses replicate rapidly and to high titer (up to 10(10) PFU/ml) in mosquito cells, producing extensive cytopathic effect and plaques, but they do not appear to replicate in mammalian cells or mice. A discussion follows on their possible biological significance and effect on mosquito vector competence for arboviruses.
Figures



References
-
- Kuno G. 2004. A survey of the relationships among the viruses not considered arboviruses, vertebrates, and arthropods. Acta Virol. 48:135–143 - PubMed
-
- Hoshino K, Isawa H, Tsuda Y, Yano K, Sasaki T, Yuda M, Takasaki T, Kobayashi M, Sawabe K. 2007. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology 359:405–414 - PubMed
-
- Igarashi A, Harrap KA, Casals J, Stollar V. 1976. Morphological, biochemical, and serological studies on a viral agent (CFA) which replicates in and causes fusion of Aedes albopictus (Singh) cells. Virology 74:174–187 - PubMed
-
- Stollar V, Thomas VL. 1975. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology 64:367–377 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

