Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1
- PMID: 23256041
- PMCID: PMC3524649
- DOI: 10.7554/eLife.00090
Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1
Abstract
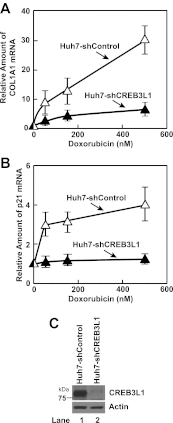
Doxorubicin is used extensively for chemotherapy of diverse types of cancer, yet the mechanism through which it inhibits proliferation of cancer cells remains unclear. Here we report that doxorubicin stimulates de novo synthesis of ceramide, which in turn activates CREB3L1, a transcription factor synthesized as a membrane-bound precursor. Doxorubicin stimulates proteolytic cleavage of CREB3L1 by Site-1 Protease and Site-2 Protease, allowing the NH(2)-terminal domain of CREB3L1 to enter the nucleus where it activates transcription of genes encoding inhibitors of the cell cycle, including p21. Knockdown of CREB3L1 mRNA in human hepatoma Huh7 cells and immortalized human fibroblast SV589 cells conferred increased resistance to doxorubicin, whereas overexpression of CREB3L1 in human breast cancer MCF-7 cells markedly enhanced the sensitivity of these cells to doxorubicin. These results suggest that measurement of CREB3L1 expression may be a useful biomarker in identifying cancer cells sensitive to doxorubicin.DOI:http://dx.doi.org/10.7554/eLife.00090.001.
Keywords: CREB3L1; Human; cancer; ceramide; doxorubicin.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
How does doxorubicin work?Elife. 2012 Dec 18;1:e00387. doi: 10.7554/eLife.00387. Elife. 2012. PMID: 23256047 Free PMC article.
References
-
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, et al. 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem 279:52772–80 - PubMed
-
- Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. 1995. Ceramide synthase mediates daunorubicin-induced apoptosis: An alternative mechanism for generating death signals. Cell 82:405–14 - PubMed
-
- Brown MS, Ye J, Rawson RB, Goldstein JL. 2000. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 100:391–8 - PubMed
-
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99:703–12 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

