In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome
- PMID: 23256155
- PMCID: PMC3545769
- DOI: 10.1073/pnas.1213624110
In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome
Abstract
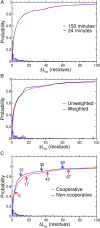
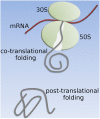
A question of fundamental importance concerning protein folding in vivo is whether the kinetics of translation or the thermodynamics of the ribosome nascent chain (RNC) complex is the major determinant of cotranslational folding behavior. This is because translation rates can reduce the probability of cotranslational folding below that associated with arrested ribosomes, whose behavior is determined by the equilibrium thermodynamics of the RNC complex. Here, we combine a chemical kinetic equation with genomic and proteomic data to predict domain folding probabilities as a function of nascent chain length for Escherichia coli cytosolic proteins synthesized on both arrested and continuously translating ribosomes. Our results indicate that, at in vivo translation rates, about one-third of the Escherichia coli cytosolic proteins exhibit cotranslational folding, with at least one domain in each of these proteins folding into its stable native structure before the full-length protein is released from the ribosome. The majority of these cotranslational folding domains are influenced by translation kinetics which reduces their probability of cotranslational folding and consequently increases the nascent chain length at which they fold into their native structures. For about 20% of all cytosolic proteins this delay in folding can exceed the length of the completely synthesized protein, causing one or more of their domains to switch from co- to posttranslational folding solely as a result of the in vivo translation rates. These kinetic effects arise from the difference in time scales of folding and amino-acid addition, and they represent a source of metastability in Escherichia coli's proteome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Nicola AV, Chen W, Helenius A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol. 1999;1(6):341–345. - PubMed
-
- Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388(6640):343–349. - PubMed
-
- Agashe VR, et al. Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell. 2004;117(2):199–209. - PubMed
-
- Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462(3):387–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

