APOBEC3G and APOBEC3F rarely co-mutate the same HIV genome
- PMID: 23256516
- PMCID: PMC3532371
- DOI: 10.1186/1742-4690-9-113
APOBEC3G and APOBEC3F rarely co-mutate the same HIV genome
Abstract
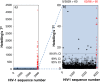
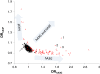
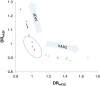
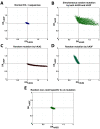
Background: The human immune proteins APOBEC3G and APOBEC3F (hA3G and hA3F) induce destructive G-to-A changes in the HIV genome, referred to as 'hypermutation'. These two proteins co-express in human cells, co-localize to mRNA processing bodies and might co-package into HIV virions. Therefore they are expected to also co-mutate the HIV genome. Here we investigate the mutational footprints of hA3G and hA3F in a large population of full genome HIV-1 sequences from naturally infected patients to uniquely identify sequences hypermutated by either or both of these proteins. We develop a method of identification based on the representation of hA3G and hA3F target and product motifs that does not require an alignment to a parental/consensus sequence.
Results: Out of nearly 100 hypermutated HIV-1 sequences only one sequence from the HIV-1 outlier group showed clear signatures of co-mutation by both proteins. The remaining sequences were affected by either hA3G or hA3F.
Conclusion: Using a novel method of identification of HIV sequences hypermutated by the hA3G and hA3F enzymes, we report a very low rate of co-mutation of full-length HIV sequences, and discuss the potential mechanisms underlying this.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

