A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme
- PMID: 23257181
- PMCID: PMC3565918
- DOI: 10.1124/pr.112.006809
A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme
Erratum in
- Pharmacol Rev. 2013;65(1):544
Abstract
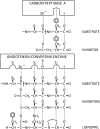
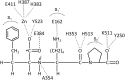
Angiotensin-converting enzyme (ACE) is a zinc-dependent peptidase responsible for converting angiotensin I into the vasoconstrictor angiotensin II. However, ACE is a relatively nonspecific peptidase that is capable of cleaving a wide range of substrates. Because of this, ACE and its peptide substrates and products affect many physiologic processes, including blood pressure control, hematopoiesis, reproduction, renal development, renal function, and the immune response. The defining feature of ACE is that it is composed of two homologous and independently catalytic domains, the result of an ancient gene duplication, and ACE-like genes are widely distributed in nature. The two ACE catalytic domains contribute to the wide substrate diversity of ACE and, by extension, the physiologic impact of the enzyme. Several studies suggest that the two catalytic domains have different biologic functions. Recently, the X-ray crystal structure of ACE has elucidated some of the structural differences between the two ACE domains. This is important now that ACE domain-specific inhibitors have been synthesized and characterized. Once widely available, these reagents will undoubtedly be powerful tools for probing the physiologic actions of each ACE domain. In turn, this knowledge should allow clinicians to envision new therapies for diseases not currently treated with ACE inhibitors.
Figures



References
-
- AbdAlla S, Lother H, Langer A, el Faramawy Y, Quitterer U. (2004) Factor XIIIA transglutaminase crosslinks AT1 receptor dimers of monocytes at the onset of atherosclerosis. Cell 119:343–354 - PubMed
-
- Agerholm-Larsen B, Nordestgaard BG, Tybjaerg-Hansen A. (2000) ACE gene polymorphism in cardiovascular disease: meta-analyses of small and large studies in whites. Arterioscler Thromb Vasc Biol 20:484–492 - PubMed
-
- Agerholm-Larsen B, Nordestgaard BG, Steffensen R, Sørensen TI, Jensen G, Tybjaerg-Hansen A. (1997) ACE gene polymorphism: ischemic heart disease and longevity in 10,150 individuals. A case-referent and retrospective cohort study based on the Copenhagen City Heart Study. Circulation 95:2358–2367 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

