Effect of capsid confinement on the chromatin organization of the SV40 minichromosome
- PMID: 23258701
- PMCID: PMC3561987
- DOI: 10.1093/nar/gks1270
Effect of capsid confinement on the chromatin organization of the SV40 minichromosome
Abstract
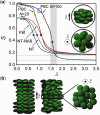
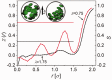
Using small-angle X-ray scattering, we determined the three-dimensional packing architecture of the minichromosome confined within the SV40 virus. In solution, the minichromosome, composed of closed circular dsDNA complexed in nucleosomes, was shown to be structurally similar to cellular chromatin. In contrast, we find a unique organization of the nanometrically encapsidated chromatin, whereby minichromosomal density is somewhat higher at the center of the capsid and decreases towards the walls. This organization is in excellent agreement with a coarse-grained computer model, accounting for tethered nucleosomal interactions under viral capsid confinement. With analogy to confined liquid crystals, but contrary to the solenoid structure of cellular chromatin, our simulations indicate that the nucleosomes within the capsid lack orientational order. Nucleosomes in the layer adjacent to the capsid wall, however, align with the boundary, thereby inducing a 'molten droplet' state of the chromatin. These findings indicate that nucleosomal interactions suffice to predict the genome organization in polyomavirus capsids and underscore the adaptable nature of the eukaryotic chromatin architecture to nanoscale confinement.
Figures


 .
.
 .
.References
-
- Cerritelli ME, Cheng NQ, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. - PubMed
-
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. - PubMed
-
- Petrov AS, Harvey SC. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

