Evolutionary dynamics of gene and isoform regulation in Mammalian tissues
- PMID: 23258891
- PMCID: PMC3568499
- DOI: 10.1126/science.1228186
Evolutionary dynamics of gene and isoform regulation in Mammalian tissues
Abstract
Most mammalian genes produce multiple distinct messenger RNAs through alternative splicing, but the extent of splicing conservation is not clear. To assess tissue-specific transcriptome variation across mammals, we sequenced complementary DNA from nine tissues from four mammals and one bird in biological triplicate, at unprecedented depth. We find that while tissue-specific gene expression programs are largely conserved, alternative splicing is well conserved in only a subset of tissues and is frequently lineage-specific. Thousands of previously unknown, lineage-specific, and conserved alternative exons were identified; widely conserved alternative exons had signatures of binding by MBNL, PTB, RBFOX, STAR, and TIA family splicing factors, implicating them as ancestral mammalian splicing regulators. Our data also indicate that alternative splicing often alters protein phosphorylatability, delimiting the scope of kinase signaling.
Figures
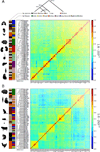
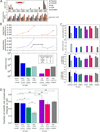


Comment in
-
Evolution. Splicing in 4D.Science. 2012 Dec 21;338(6114):1547-8. doi: 10.1126/science.1233219. Science. 2012. PMID: 23258879 No abstract available.
References
-
- Stamm S, et al. Gene. 2005 Jan 3;344:1. - PubMed
-
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. Advances in experimental medicine and biology. 2007;623:190. - PubMed
-
- Wang ET, et al. Nature. 2008 Nov 27;456:470. - PubMed
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Nature genetics. 2008 Dec;40:1413. - PubMed
-
- Brawand D, et al. Nature. 2011 Oct 20;478:343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

