Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing
- PMID: 23258895
- PMCID: PMC3590491
- DOI: 10.1126/science.1229112
Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing
Abstract
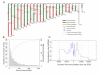
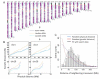
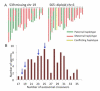
Meiotic recombination creates genetic diversity and ensures segregation of homologous chromosomes. Previous population analyses yielded results averaged among individuals and affected by evolutionary pressures. We sequenced 99 sperm from an Asian male by using the newly developed amplification method-multiple annealing and looping-based amplification cycles-to phase the personal genome and map recombination events at high resolution, which are nonuniformly distributed across the genome in the absence of selection pressure. The paucity of recombination near transcription start sites observed in individual sperm indicates that such a phenomenon is intrinsic to the molecular mechanism of meiosis. Interestingly, a decreased crossover frequency combined with an increase of autosomal aneuploidy is observable on a global per-sperm basis.
Figures

Comment in
-
Single cells go fully genomic.Nat Methods. 2013 Mar;10(3):190-1. doi: 10.1038/nmeth.2391. Nat Methods. 2013. PMID: 23570041 No abstract available.
References
-
- Petronczki M, Siomos MF, Nasmyth K. Un Ménage à Quatre: The Molecular Biology of Chromosome Segregation in Meiosis. Cell. 2003;112:423–440. - PubMed
-
- Epstein CJ. The consequences of chromosome imbalance: principles, mechanisms, and models. Cambridge Univ Pr; 2007.
-
- Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nature Genetics. 2004;36:151–156. - PubMed
-
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A Fine-Scale Map of Recombination Rates and Hotspots Across the Human Genome. Science. 2005;310:321–324. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

