Influence of a Gamma Amino Acid on the Structures and Reactivity of Peptide a(3) Ions
- PMID: 23258959
- PMCID: PMC3523335
- DOI: 10.1016/j.ijms.2012.02.028
Influence of a Gamma Amino Acid on the Structures and Reactivity of Peptide a(3) Ions
Abstract
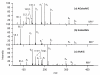
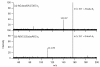
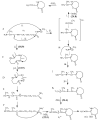
Collision-induced dissociation of protonated AGabaAIG (where Gaba is gamma-amino butyric acid, NH(2)-(CH(2))(3)-COOH) leads to an unusually stable a(3) ion. Tandem mass spectrometry and theory are used here to probe the enhanced stability of this fragment, whose counterpart is not usually observed in CID of protonated peptides containing only alpha amino acids. Experiments are carried out on the unlabelled and (15)N-Ala labeled AGabaAIG (labeled separately at residue one or three) probing the b(3), a(3), a(3)-NH(3) (a(3) (*)), and b(2) fragments while theory is used to characterize the most stable b(3), a(3), and b(2) structures and the formation and dissociation of the a(3) ion. Our results indicate the AGabaA oxazolone b(3) isomer undergoes head-to-tail macrocyclization and subsequent ring opening to form the GabaAA sequence isomer while this chemistry is energetically disfavored for the AAA sequence. The AGabaA a(3) fragment also undergoes macrocyclization and rearrangement to form the rearranged imine-amide isomer while this reaction is energetically disfavored for the AAA sequence. The barriers to dissociation of the AGabaA a(3) ion via the a(3)→b(2) and a(3)→a(3)* channels are higher than the literature values reported for the AAA sequence. These two effects provide a clear explanation for the enhanced stability of the AGabaA a(3) ion.
Figures



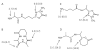




Similar articles
-
Cyclization and rearrangement reactions of a(n) fragment ions of protonated peptides.J Am Chem Soc. 2010 Oct 27;132(42):14766-79. doi: 10.1021/ja101556g. J Am Chem Soc. 2010. PMID: 20925356
-
Sequence-scrambling fragmentation pathways of protonated peptides.J Am Chem Soc. 2008 Dec 31;130(52):17774-89. doi: 10.1021/ja805074d. J Am Chem Soc. 2008. PMID: 19055406
-
Specific rearrangement reactions of acetylated lysine containing peptide bn (n = 4-7) ion series.J Mass Spectrom. 2014 Dec;49(12):1290-7. doi: 10.1002/jms.3462. J Mass Spectrom. 2014. PMID: 25476947
-
Electrospray ionization tandem mass spectrometric study of protonated and alkali- cationized α/ε-hybrid peptides: differentiation of a pair of dipeptide positional isomers.Eur J Mass Spectrom (Chichester). 2016;22(4):181-191. doi: 10.1255/ejms.1431. Eur J Mass Spectrom (Chichester). 2016. PMID: 27882883
-
Effect of peptide fragment size on the propensity of cyclization in collision-induced dissociation: oligoglycine b(2)-b(8).J Am Chem Soc. 2009 Dec 30;131(51):18272-82. doi: 10.1021/ja9030837. J Am Chem Soc. 2009. PMID: 19947633
Cited by
-
Spectroscopic Evidence for Lactam Formation in Terminal Ornithine b2+ and b3+ Fragment Ions.J Am Soc Mass Spectrom. 2019 Sep;30(9):1565-1577. doi: 10.1007/s13361-019-02244-0. Epub 2019 Jun 10. J Am Soc Mass Spectrom. 2019. PMID: 31183839 Free PMC article.
References
-
- Paizs B, Suhai S. Mass Spec. Rev. 2005;24(4):508–548. - PubMed
-
- Breci LA, Tabb DL, Yates JR, III, Wysocki VH. Anal. Chem. 2003;75(9):1963–1971. - PubMed
-
- Roepstorff P, Fohlmann J. Proposals for a Common Nomenclature for Sequence Ions in Mass Spectra of Peptides. Biomed. Mass Spectrom. 1984;11:601. - PubMed
-
- Biemann K. Contributions of Mass Spectrometry to Peptide and Protein Structure. Biomed. Env. Mass Spectrom. 1988;16:99–111. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
