Cascade of reduced speed and accuracy after errors in enzyme-free copying of nucleic acid sequences
- PMID: 23259600
- PMCID: PMC3557965
- DOI: 10.1021/ja3095558
Cascade of reduced speed and accuracy after errors in enzyme-free copying of nucleic acid sequences
Abstract
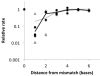
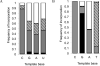
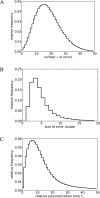
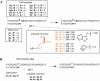
Nonenzymatic, template-directed synthesis of nucleic acids is a paradigm for self-replicating systems. The evolutionary dynamics of such systems depend on several factors, including the mutation rates, relative replication rates, and sequence characteristics of mutant sequences. We measured the kinetics of correct and incorrect monomer insertion downstream of a primer-template mismatch (mutation), using a range of backbone structures (RNA, DNA, and LNA templates and RNA and DNA primers) and two types of 5'-activated nucleotides (oxyazabenzotriazolides and imidazolides, i.e., nucleoside 5'-phosphorimidazolides). Our study indicated that for all systems studied, an initial mismatch was likely to be followed by another error (54-75% of the time), and extension after a single mismatch was generally 10-100 times slower than extension without errors. If the mismatch was followed by a matched base pair, the extension rate recovered to nearly normal levels. On the basis of these data, we simulated nucleic acid replication in silico, which indicated that a primer suffering an initial error would lag behind properly extended counterparts due to a cascade of subsequent errors and kinetic stalling, with the typical mutational event consisting of several consecutive errors. Our study also included different sequence contexts, which suggest the presence of cooperativity among monomers affecting both absolute rate (by up to 2 orders of magnitude) and fidelity. The results suggest that molecular evolution in enzyme-free replication systems would be characterized by large "leaps" through sequence space rather than isolated point mutations, perhaps enabling rapid exploration of diverse sequences. The findings may also be useful for designing self-replicating systems combining high fidelity with evolvability.
Figures
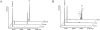
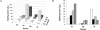
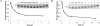



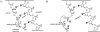
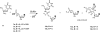
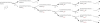
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

