HLA typing from RNA-Seq sequence reads
- PMID: 23259685
- PMCID: PMC4064318
- DOI: 10.1186/gm403
HLA typing from RNA-Seq sequence reads
Abstract
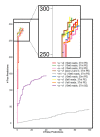
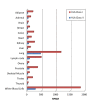
We present a method, seq2HLA, for obtaining an individual's human leukocyte antigen (HLA) class I and II type and expression using standard next generation sequencing RNA-Seq data. RNA-Seq reads are mapped against a reference database of HLA alleles, and HLA type, confidence score and locus-specific expression level are determined. We successfully applied seq2HLA to 50 individuals included in the HapMap project, yielding 100% specificity and 94% sensitivity at a P-value of 0.1 for two-digit HLA types. We determined HLA type and expression for previously un-typed Illumina Body Map tissues and a cohort of Korean patients with lung cancer. Because the algorithm uses standard RNA-Seq reads and requires no change to laboratory protocols, it can be used for both existing datasets and future studies, thus adding a new dimension for HLA typing and biomarker studies.
Figures


References
-
- Cao K, Chopek M, Fernandez-Vina MA. High and intermediate resolution DNA typing systems for class I HLA-A, B, C genes by hybridization with sequence-specific oligonucleotide probes (SSOP) Rev Immunogenet. 1999;4:177–208. - PubMed
-
- Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;4:225–235. doi: 10.1111/j.1399-0039.1992.tb01940.x. - DOI - PubMed
-
- Terasaki PI, McClelland JD. Microdroplet assay of human serum cytotoxins. Nature. 1964;4:998–1000. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

