HIV-1 integrase resistance among antiretroviral treatment naive and experienced patients from Northwestern Poland
- PMID: 23259737
- PMCID: PMC3547692
- DOI: 10.1186/1471-2334-12-368
HIV-1 integrase resistance among antiretroviral treatment naive and experienced patients from Northwestern Poland
Abstract
Background: HIV integrase inhibitor use is limited by low genetic barrier to resistance and possible cross-resistance among representatives of this class of antiretrovirals. The aim of this study was to analyse integrase sequence variability among antiretroviral treatment naive and experienced patients with no prior integrase inhibitor (InI) exposure and investigate development of the InI drug resistance mutations following the virologic failure of the raltegravir containing regimen.
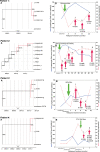
Methods: Sequencing of HIV-1 integrase region from plasma samples of 80 integrase treatment naive patients and serial samples from 12 patients with observed virologic failure on raltegravir containing treatment whenever plasma vireamia exceeded >50 copies/ml was performed. Drug resistance mutations were called with Stanford DB database and grouped into major and minor variants. For subtyping bootstrapped phylogenetic analysis was used; Bayesian Monte Carlo Marcov Chain (MCMC) model was implemented to infer on the phylogenetic relationships between the serial sequences from patients failing on raltegravir.
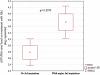
Results: Majority of the integrase region sequences were classified as subtype B; the remaining ones being subtype D, C, G, as well as CRF01_AE , CRF02_AG and CRF13_cpx recombinants. No major integrase drug resistance mutations have been observed in InI-treatment naive patients. In 30 (38.5%) cases polymorphic variation with predominance of the E157Q mutation was observed. This mutation was more common among subtype B (26 cases, 54.2%) than non-B sequences (5 cases, 16.7%), p=0.00099, OR: 5.91 (95% CI:1.77-22.63)]. Other variants included L68V, L74IL, T97A, E138D, V151I, R263K. Among 12 (26.1%) raltegravir treated patients treatment failure was observed; major InI drug resistance mutations (G140S, Q148H and N155H, V151I, E92EQ, V151I, G163R) were noted in four of these cases (8.3% of the total InI-treated patients). Time to the development of drug resistance ranged from 2.6 to 16.3 months with mean increase of HIV viral load of 4.34 (95% CI:1.86-6.84) log HIV-RNA copies/ml at the time of emergence of the major mutations. Baseline polymorphisms, including E157Q were not associated with the virologic failure on raltegravir.
Conclusions: In InI treatment naive patients polymorphic integrase sequence variation was common, with no major resistance mutants. In the treatment failing patients selection of drug resistance occurred rapidly and followed the typical drug resistance pathways. Preexisting integrase polymorphisms were not associated with the treatment failure.
Figures

Similar articles
-
Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades.J Antimicrob Chemother. 2015 Nov;70(11):3080-6. doi: 10.1093/jac/dkv243. Epub 2015 Aug 26. J Antimicrob Chemother. 2015. PMID: 26311843 Free PMC article.
-
HIV-1 integrase sequence variability in antiretroviral naïve patients and in triple-class experienced patients subsequently treated with raltegravir.AIDS Res Hum Retroviruses. 2010 Dec;26(12):1323-6. doi: 10.1089/aid.2010.0123. Epub 2010 Oct 21. AIDS Res Hum Retroviruses. 2010. PMID: 20961278 Free PMC article.
-
HIV type 1 integrase polymorphisms in treatment-naive and treatment-experienced HIV type 1-infected patients in Thailand where HIV type 1 subtype A/E predominates.AIDS Res Hum Retroviruses. 2012 Aug;28(8):937-43. doi: 10.1089/AID.2011.0139. Epub 2011 Nov 22. AIDS Res Hum Retroviruses. 2012. PMID: 21970343
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
Resistance to HIV Integrase Inhibitors: About R263K and E157Q Mutations.Viruses. 2018 Jan 18;10(1):41. doi: 10.3390/v10010041. Viruses. 2018. PMID: 29346270 Free PMC article. Review.
Cited by
-
Prevalence of resistance mutations associated with integrase inhibitors in therapy-naive HIV-positive patients in Baoding, Hebei province, China.Front Genet. 2022 Sep 14;13:975397. doi: 10.3389/fgene.2022.975397. eCollection 2022. Front Genet. 2022. PMID: 36186451 Free PMC article.
-
Effect of HIV-1 Subtype C integrase mutations implied using molecular modeling and docking data.Bioinformation. 2016 Jun 15;12(3):221-230. doi: 10.6026/97320630012221. eCollection 2016. Bioinformation. 2016. PMID: 28149058 Free PMC article.
-
Low prevalence of archived integrase strand transfer inhibitors resistance associated mutations in Botswana before the roll out of dolutegravir based first line antiretroviral therapy.Front Microbiol. 2024 Oct 24;15:1482348. doi: 10.3389/fmicb.2024.1482348. eCollection 2024. Front Microbiol. 2024. PMID: 39512940 Free PMC article.
-
Virologic failure after 48 weeks of raltegravir-based regimen in low HIV-1 incidence setting.Antivir Chem Chemother. 2020 Jan-Dec;28:2040206620927908. doi: 10.1177/2040206620927908. Antivir Chem Chemother. 2020. PMID: 32434393 Free PMC article.
-
CODEHOP-Mediated PCR Improves HIV-1 Genotyping and Detection of Variants by MinION Sequencing.Microbiol Spectr. 2021 Oct 31;9(2):e0143221. doi: 10.1128/Spectrum.01432-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668751 Free PMC article.
References
-
- Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, Zhao J, Xu X, Williams-Diaz A, Rodgers AJ. et al.Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet. 2009;374(9692):796–806. doi: 10.1016/S0140-6736(09)60918-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

