Protein interface classification by evolutionary analysis
- PMID: 23259833
- PMCID: PMC3556496
- DOI: 10.1186/1471-2105-13-334
Protein interface classification by evolutionary analysis
Abstract
Background: Distinguishing biologically relevant interfaces from lattice contacts in protein crystals is a fundamental problem in structural biology. Despite efforts towards the computational prediction of interface character, many issues are still unresolved.
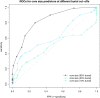
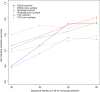
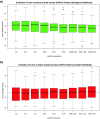
Results: We present here a protein-protein interface classifier that relies on evolutionary data to detect the biological character of interfaces. The classifier uses a simple geometric measure, number of core residues, and two evolutionary indicators based on the sequence entropy of homolog sequences. Both aim at detecting differential selection pressure between interface core and rim or rest of surface. The core residues, defined as fully buried residues (>95% burial), appear to be fundamental determinants of biological interfaces: their number is in itself a powerful discriminator of interface character and together with the evolutionary measures it is able to clearly distinguish evolved biological contacts from crystal ones. We demonstrate that this definition of core residues leads to distinctively better results than earlier definitions from the literature. The stringent selection and quality filtering of structural and sequence data was key to the success of the method. Most importantly we demonstrate that a more conservative selection of homolog sequences - with relatively high sequence identities to the query - is able to produce a clearer signal than previous attempts.
Conclusions: An evolutionary approach like the one presented here is key to the advancement of the field, which so far was missing an effective method exploiting the evolutionary character of protein interfaces. Its coverage and performance will only improve over time thanks to the incessant growth of sequence databases. Currently our method reaches an accuracy of 89% in classifying interfaces of the Ponstingl 2003 datasets and it lends itself to a variety of useful applications in structural biology and bioinformatics. We made the corresponding software implementation available to the community as an easy-to-use graphical web interface at http://www.eppic-web.org.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

