pH tuning of DNA translocation time through organically functionalized nanopores
- PMID: 23259840
- PMCID: PMC3584232
- DOI: 10.1021/nn3051677
pH tuning of DNA translocation time through organically functionalized nanopores
Abstract
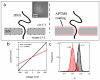
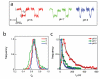
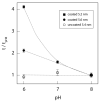
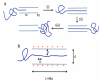
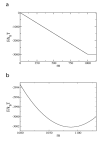
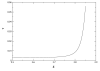
Controlling DNA translocation speed is critically important for nanopore sequencing as free electrophoretic threading is far too rapid to resolve individual bases. A number of promising strategies have been explored in recent years, largely driven by the demands of next-generation sequencing. Engineering DNA-nanopore interactions (known to dominate translocation dynamics) with organic coatings is an attractive method as it does not require sample modification, processive enzymes, or complicated and expensive fabrication steps. In this work, we show for the first time 4-fold tuning of unfolded, single-file translocation time through small, amine-functionalized solid-state nanopores by varying the solution pH in situ. Additionally, we develop a simple analytical model based on electrostatic interactions to explain this effect which will be a useful tool in designing future devices and experiments.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

