Novel polyomavirus associated with brain tumors in free-ranging raccoons, western United States
- PMID: 23260029
- PMCID: PMC3558004
- DOI: 10.3201/eid1901.121078
Novel polyomavirus associated with brain tumors in free-ranging raccoons, western United States
Abstract
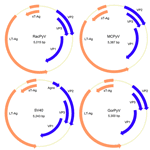
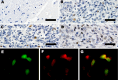
Tumors of any type are exceedingly rare in raccoons. High-grade brain tumors, consistently located in the frontal lobes and olfactory tracts, were detected in 10 raccoons during March 2010-May 2012 in California and Oregon, suggesting an emerging, infectious origin. We have identified a candidate etiologic agent, dubbed raccoon polyomavirus, that was present in the tumor tissue of all affected animals but not in tissues from 20 unaffected animals. Southern blot hybridization and rolling circle amplification showed the episomal viral genome in the tumors. The multifunctional nuclear protein large T-antigen was detectable by immunohistochemical analyses in a subset of neoplastic cells. Raccoon polyomavirus may contribute to the development of malignant brain tumors of raccoons.
Figures




Similar articles
-
UC-Davis veterinarians identify new raccoon polyomavirus: discovery could help explain how viruses cause cancer in animals and humans.J Am Vet Med Assoc. 2013 Feb 1;242(3):295-6. J Am Vet Med Assoc. 2013. PMID: 23447820 No abstract available.
-
Polyomavirus and Naturally Occuring Neuroglial Tumors in Raccoons (Procyon Lotor).ILAR J. 2016;56(3):297-305. doi: 10.1093/ilar/ilv036. ILAR J. 2016. PMID: 26912716
-
Temporal and geographic clustering of polyomavirus-associated olfactory tumors in 10 free-ranging raccoons (Procyon lotor).Vet Pathol. 2014 Jul;51(4):832-45. doi: 10.1177/0300985813502817. Epub 2013 Sep 17. Vet Pathol. 2014. PMID: 24045888
-
From Stockholm to Malawi: recent developments in studying human polyomaviruses.J Gen Virol. 2013 Mar;94(Pt 3):482-496. doi: 10.1099/vir.0.048462-0. Epub 2012 Dec 19. J Gen Virol. 2013. PMID: 23255626 Review.
-
Polyomaviruses of nonhuman primates: implications for research.Comp Med. 2008 Feb;58(1):51-6. Comp Med. 2008. PMID: 19793457 Free PMC article. Review.
Cited by
-
Prospective investigation of polyomavirus infection and the risk of adult glioma.Sci Rep. 2021 May 5;11(1):9642. doi: 10.1038/s41598-021-89133-3. Sci Rep. 2021. PMID: 33953301 Free PMC article.
-
Polyomavirus surveillance in cetaceans of Brazil: first detection of polyomavirus in Guiana dolphins (Sotalia guianensis).Vet Q. 2024 Dec;44(1):1-7. doi: 10.1080/01652176.2024.2413185. Epub 2024 Oct 20. Vet Q. 2024. PMID: 39428385 Free PMC article.
-
Recent advances in discovery and functional analysis of the small proteins and microRNA expressed by polyomaviruses.Virology. 2025 Jan;602:110310. doi: 10.1016/j.virol.2024.110310. Epub 2024 Nov 22. Virology. 2025. PMID: 39612622 Free PMC article. Review.
-
Extensive Genetic Diversity of Polyomaviruses in Sympatric Bat Communities: Host Switching versus Coevolution.J Virol. 2020 Apr 16;94(9):e02101-19. doi: 10.1128/JVI.02101-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32075934 Free PMC article.
-
Discovery of a polyomavirus in European badgers (Meles meles) and the evolution of host range in the family Polyomaviridae.J Gen Virol. 2015 Jun;96(Pt 6):1411-1422. doi: 10.1099/vir.0.000071. Epub 2015 Jan 27. J Gen Virol. 2015. PMID: 25626684 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
