Role of nonspecific interactions in molecular chaperones through model-based bioinformatics
- PMID: 23260050
- PMCID: PMC3525852
- DOI: 10.1016/j.bpj.2012.10.040
Role of nonspecific interactions in molecular chaperones through model-based bioinformatics
Abstract
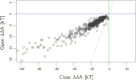
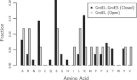
Molecular chaperones are large proteins or protein complexes from which many proteins require assistance in order to fold. One unique property of molecular chaperones is the cavity they provide in which proteins fold. The interior surface residues which make up the cavities of molecular chaperone complexes from different organisms has recently been identified, including the well-studied GroEL-GroES chaperonin complex found in Escherichia coli. It was found that the interior of these protein complexes is significantly different than other protein surfaces and that the residues found on the protein surface are able to resist protein adsorption when immobilized on a surface. Yet it remains unknown if these residues passively resist protein binding inside GroEL-GroEs (as demonstrated by experiments that created synthetic mimics of the interior cavity) or if the interior also actively stabilizes protein folding. To answer this question, we have extended entropic models of substrate protein folding inside GroEL-GroES to include interaction energies between substrate proteins and the GroEL-GroES chaperone complex. This model was tested on a set of 528 proteins and the results qualitatively match experimental observations. The interior residues were found to strongly discourage the exposure of any hydrophobic residues, providing an enhanced hydrophobic effect inside the cavity that actively influences protein folding. This work provides both a mechanism for active protein stabilization in GroEL-GroES and a model that matches contemporary understanding of the chaperone protein.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The 69 kDa Escherichia coli maltodextrin glucosidase does not get encapsulated underneath GroES and folds through trans mechanism during GroEL/GroES-assisted folding.FASEB J. 2007 Sep;21(11):2874-85. doi: 10.1096/fj.06-7958com. Epub 2007 May 10. FASEB J. 2007. PMID: 17494995
-
The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding.Trends Biochem Sci. 2016 Jan;41(1):62-76. doi: 10.1016/j.tibs.2015.07.009. Epub 2015 Sep 25. Trends Biochem Sci. 2016. PMID: 26422689 Review.
-
Productive folding of a tethered protein in the chaperonin GroEL-GroES cage.Biochem Biophys Res Commun. 2015 Oct 9;466(1):72-5. doi: 10.1016/j.bbrc.2015.08.108. Epub 2015 Aug 29. Biochem Biophys Res Commun. 2015. PMID: 26325470
-
Co-expression of chaperonin GroEL/GroES enhances in vivo folding of yeast mitochondrial aconitase and alters the growth characteristics of Escherichia coli.Int J Biochem Cell Biol. 2006;38(11):1975-85. doi: 10.1016/j.biocel.2006.05.013. Epub 2006 Jun 2. Int J Biochem Cell Biol. 2006. PMID: 16822698
-
Molecular chaperone GroEL/ES: unfolding and refolding processes.Biochemistry (Mosc). 2013 Dec;78(13):1405-14. doi: 10.1134/S0006297913130038. Biochemistry (Mosc). 2013. PMID: 24490731 Review.
Cited by
-
Learning peptide properties with positive examples only.Digit Discov. 2024 Apr 19;3(5):977-986. doi: 10.1039/d3dd00218g. eCollection 2024 May 15. Digit Discov. 2024. PMID: 38756224 Free PMC article.
-
Serverless Prediction of Peptide Properties with Recurrent Neural Networks.J Chem Inf Model. 2023 Apr 24;63(8):2546-2553. doi: 10.1021/acs.jcim.2c01317. Epub 2023 Apr 3. J Chem Inf Model. 2023. PMID: 37010950 Free PMC article.
-
PeptideBERT: A Language Model Based on Transformers for Peptide Property Prediction.J Phys Chem Lett. 2023 Nov 23;14(46):10427-10434. doi: 10.1021/acs.jpclett.3c02398. Epub 2023 Nov 13. J Phys Chem Lett. 2023. PMID: 37956397 Free PMC article.
-
Effects of 4-Hexylresorcinol on Protein Expressions in RAW 264.7 Cells as Determined by Immunoprecipitation High Performance Liquid Chromatography.Sci Rep. 2019 Mar 4;9(1):3379. doi: 10.1038/s41598-019-38946-4. Sci Rep. 2019. PMID: 30833641 Free PMC article.
References
-
- Onuchic J.N., Luthey-Schulten Z., Wolynes P.G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 1997;48:545–600. - PubMed
-
- Yancey P.H., Clark M.E., Somero G.N. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. - PubMed
-
- Horwich A.L., Fenton W.A., Farr G.W. Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 2007;23:115–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

