Kinetics of the triplex-duplex transition in DNA
- PMID: 23260051
- PMCID: PMC3525853
- DOI: 10.1016/j.bpj.2012.10.029
Kinetics of the triplex-duplex transition in DNA
Abstract
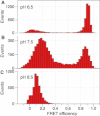
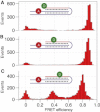
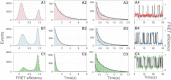
The kinetics of triplex folding/unfolding is investigated by the single-molecule fluorescence resonance energy transfer (FRET) technique. In neutral pH conditions, the average dwell times in both high-FRET (folded) and low-FRET (unfolded) states are comparable, meaning that the triplex is marginally stable. The dwell-time distributions are qualitatively different: while the dwell-time distribution of the high-FRET state should be fit with at least a double-exponential function, the dwell-time distribution of the low-FRET state can be fit with a single-exponential function. We propose a model where the folding can be trapped in metastable states, which is consistent with the FRET data. Our model also accounts for the fact that the relevant timescales of triplex folding/unfolding are macroscopic.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
DNA triplex folding: moderate versus high salt conditions.Eur Phys J E Soft Matter. 2013 Jun;36(6):57. doi: 10.1140/epje/i2013-13057-4. Epub 2013 Jun 10. Eur Phys J E Soft Matter. 2013. PMID: 23749234
-
Folding and unfolding pathways of the human telomeric G-quadruplex.J Mol Biol. 2014 Apr 17;426(8):1629-50. doi: 10.1016/j.jmb.2014.01.009. Epub 2014 Jan 31. J Mol Biol. 2014. PMID: 24487181 Free PMC article.
-
Fluorescence-Detected Conformational Changes in Duplex DNA in Open Complex Formation by Escherichia coli RNA Polymerase: Upstream Wrapping and Downstream Bending Precede Clamp Opening and Insertion of the Downstream Duplex.Biochemistry. 2020 Apr 28;59(16):1565-1581. doi: 10.1021/acs.biochem.0c00098. Epub 2020 Apr 7. Biochemistry. 2020. PMID: 32216369 Free PMC article.
-
New approaches toward recognition of nucleic acid triple helices.Acc Chem Res. 2011 Feb 15;44(2):134-46. doi: 10.1021/ar100113q. Epub 2010 Nov 12. Acc Chem Res. 2011. PMID: 21073199 Free PMC article. Review.
-
Using sm-FRET and denaturants to reveal folding landscapes.Methods Enzymol. 2014;549:313-41. doi: 10.1016/B978-0-12-801122-5.00014-3. Methods Enzymol. 2014. PMID: 25432755 Review.
Cited by
-
DNA triplex folding: moderate versus high salt conditions.Eur Phys J E Soft Matter. 2013 Jun;36(6):57. doi: 10.1140/epje/i2013-13057-4. Epub 2013 Jun 10. Eur Phys J E Soft Matter. 2013. PMID: 23749234
-
Cation-induced kinetic heterogeneity of the intron-exon recognition in single group II introns.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3403-8. doi: 10.1073/pnas.1322759112. Epub 2015 Mar 3. Proc Natl Acad Sci U S A. 2015. PMID: 25737541 Free PMC article.
-
Decoding Single Molecule Time Traces with Dynamic Disorder.PLoS Comput Biol. 2016 Dec 27;12(12):e1005286. doi: 10.1371/journal.pcbi.1005286. eCollection 2016 Dec. PLoS Comput Biol. 2016. PMID: 28027304 Free PMC article.
-
First passage time study of DNA strand displacement.Biophys J. 2021 Jun 15;120(12):2400-2412. doi: 10.1016/j.bpj.2021.01.043. Epub 2021 Apr 22. Biophys J. 2021. PMID: 33894217 Free PMC article.
-
NALDB: nucleic acid ligand database for small molecules targeting nucleic acid.Database (Oxford). 2016 Feb 20;2016:baw002. doi: 10.1093/database/baw002. Print 2016. Database (Oxford). 2016. PMID: 26896846 Free PMC article.
References
-
- Felsenfeld G., Davies D.R., Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024.
-
- Frank-Kamenetskii M.D., Mirkin S.M. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. - PubMed
-
- Soyfer V., Potaman V.N. Springer; New York: 1996. Triple-Helical Nucleic Acids.
-
- Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1963;16:907–916.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

