Limitations of time-resolved fluorescence suggested by molecular simulations: assessing the dynamics of T cell receptor binding loops
- PMID: 23260055
- PMCID: PMC3525847
- DOI: 10.1016/j.bpj.2012.10.037
Limitations of time-resolved fluorescence suggested by molecular simulations: assessing the dynamics of T cell receptor binding loops
Abstract
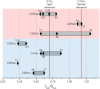
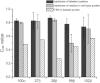
Time-resolved fluorescence anisotropy (TRFA) has a rich history in evaluating protein dynamics. Yet as often employed, TRFA assumes that the motional properties of a covalently tethered fluorescent probe accurately portray the motional properties of the protein backbone at the probe attachment site. In an extensive survey using TRFA to study the dynamics of the binding loops of a αβ T cell receptor, we observed multiple discrepancies between the TRFA data and previously published results that led us to question this assumption. We thus simulated several of the experimentally probed systems using a protocol that permitted accurate determination of probe and protein time correlation functions. We found excellent agreement in the decays of the experimental and simulated correlation functions. However, the motional properties of the probe were poorly correlated with those of the backbone of both the labeled and unlabeled protein. Our results warrant caution in the interpretation of TRFA data and suggest further studies to ascertain the extent to which probe dynamics reflect those of the protein backbone. Meanwhile, the agreement between experiment and computation validates the use of molecular dynamics simulations as an accurate tool for exploring the molecular motion of T cell receptors and their binding loops.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Boyd A.E., Dunlop C.S., Johnson D.A. Nanosecond dynamics of acetylcholinesterase near the active center gorge. J. Biol. Chem. 2004;279:26612–26618. - PubMed
-
- Hibbs R.E., Johnson D.A., Taylor P. Structural dynamics of the alpha-neurotoxin-acetylcholine-binding protein complex: hydrodynamic and fluorescence anisotropy decay analyses. Biochemistry. 2005;44:16602–16611. - PubMed
-
- Hibbs R.E., Radic Z., Johnson D.A. Influence of agonists and antagonists on the segmental motion of residues near the agonist binding pocket of the acetylcholine-binding protein. J. Biol. Chem. 2006;281:39708–39718. - PubMed
-
- Kim T.-Y., Schlieter T., Alexiev U. Activation and molecular recognition of the GPCR rhodopsin - insights from time-resolved fluorescence depolarisation and single molecule experiments. Eur. J. Cell Biol. 2012;91:300–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

