Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation
- PMID: 23260146
- PMCID: PMC3530164
- DOI: 10.1016/j.cell.2012.11.018
Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation
Abstract
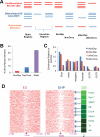
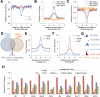
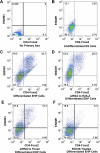
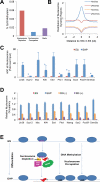
Nucleosome occupancy is fundamental for establishing chromatin architecture. However, little is known about the relationship between nucleosome dynamics and initial cell lineage specification. Here, we determine the mechanisms that control global nucleosome dynamics during embryonic stem (ES) cell differentiation into endoderm. Both nucleosome depletion and de novo occupation occur during the differentiation process, with higher overall nucleosome density after differentiation. The variant histone H2A.Z and the winged helix transcription factor Foxa2 both act to regulate nucleosome depletion and gene activation, thus promoting ES cell differentiation, whereas DNA methylation promotes nucleosome occupation and suppresses gene expression. Nucleosome depletion during ES cell differentiation is dependent on Nap1l1-coupled SWI/SNF and INO80 chromatin remodeling complexes. Thus, both epigenetic and genetic regulators cooperate to control nucleosome dynamics during ES cell fate decisions.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Comment in
-
H2A.Z sets the stage in ESCs.Cell Stem Cell. 2013 Feb 7;12(2):143-4. doi: 10.1016/j.stem.2013.01.012. Cell Stem Cell. 2013. PMID: 23395439
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

