H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation
- PMID: 23260488
- PMCID: PMC3570599
- DOI: 10.1016/j.stem.2012.11.003
H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation
Abstract
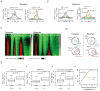
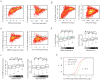
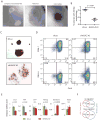
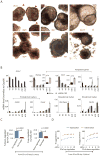
Chromatin modifications have been implicated in the self-renewal and differentiation of embryonic stem cells (ESCs). However, the function of histone variant H2A.Z in ESCs remains unclear. We show that H2A.Z is highly enriched at promoters and enhancers and is required for both efficient self-renewal and differentiation of murine ESCs. H2A.Z deposition leads to an abnormal nucleosome structure, decreased nucleosome occupancy, and increased chromatin accessibility. In self-renewing ESCs, knockdown of H2A.Z compromises OCT4 binding to its target genes and leads to decreased binding of MLL complexes to active genes and of PRC2 complex to repressed genes. During differentiation of ESCs, inhibition of H2A.Z also compromises RA-induced RARα binding, activation of differentiation markers, and the repression of pluripotency genes. We propose that H2A.Z mediates such contrasting activities by acting as a general facilitator that generates access for a variety of complexes, both activating and repressive.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures


Comment in
-
H2A.Z sets the stage in ESCs.Cell Stem Cell. 2013 Feb 7;12(2):143-4. doi: 10.1016/j.stem.2013.01.012. Cell Stem Cell. 2013. PMID: 23395439
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

