Coordinated induction of multi-gene pathways in Saccharomyces cerevisiae
- PMID: 23262224
- PMCID: PMC3581276
- DOI: 10.1093/nar/gks1293
Coordinated induction of multi-gene pathways in Saccharomyces cerevisiae
Abstract
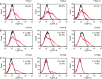
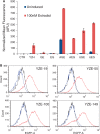
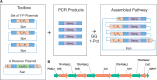
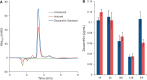
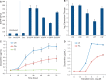
Bacterial operons are nature's tool for regulating and coordinating multi-gene expression in prokaryotes. They are also a gene architecture commonly used in the biosynthesis of many pharmaceutically important compounds and industrially useful chemicals. Despite being an important eukaryotic production host, Saccharomyces cerevisiae has never had such gene architecture. Here, we report the development of a system to assemble and regulate a multi-gene pathway in S. cerevisiae. Full pathways can be constructed using pre-made parts from a plasmid toolbox. Subsequently, through the use of a yeast strain containing a stably integrated gene switch, the assembled pathway can be regulated using a readily available and inexpensive compound-estradiol-with extremely high sensitivity (10 nM). To demonstrate the use of the system, we assembled the five-gene zeaxanthin biosynthetic pathway in a single step and showed the ligand-dependent coordinated expression of all five genes as well as the tightly regulated production of zeaxanthin. Compared with a previously reported constitutive zeaxanthin pathway, our inducible pathway was shown to have 50-fold higher production level.
Figures
References
-
- Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

