Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding
- PMID: 23262490
- PMCID: PMC3565066
- DOI: 10.1038/nsmb.2466
Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding
Abstract
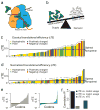
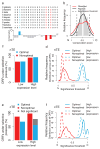
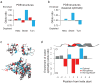
The choice of codons can influence local translation kinetics during protein synthesis. Whether codon preference is linked to cotranslational regulation of polypeptide folding remains unclear. Here, we derive a revised translational efficiency scale that incorporates the competition between tRNA supply and demand. Applying this scale to ten closely related yeast species, we uncover the evolutionary conservation of codon optimality in eukaryotes. This analysis reveals universal patterns of conserved optimal and nonoptimal codons, often in clusters, which associate with the secondary structure of the translated polypeptides independent of the levels of expression. Our analysis suggests an evolved function for codon optimality in regulating the rhythm of elongation to facilitate cotranslational polypeptide folding, beyond its previously proposed role of adapting to the cost of expression. These findings establish how mRNA sequences are generally under selection to optimize the cotranslational folding of corresponding polypeptides.
Figures



References
-
- Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

