The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion
- PMID: 23262491
- PMCID: PMC3565103
- DOI: 10.1038/nsmb.2472
The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion
Abstract
Gene expression, DNA replication and genome maintenance are all initiated by proteins that must recognize specific targets from among a vast excess of nonspecific DNA. For example, to initiate transcription, Escherichia coli RNA polymerase (RNAP) must locate promoter sequences, which compose <2% of the bacterial genome. This search problem remains one of the least understood aspects of gene expression, largely owing to the transient nature of search intermediates. Here we visualize RNAP in real time as it searches for promoters, and we develop a theoretical framework for analyzing target searches at the submicroscopic scale on the basis of single-molecule target-association rates. We demonstrate that, contrary to long-held assumptions, the promoter search is dominated by three-dimensional diffusion at both the microscopic and submicroscopic scales in vitro, which has direct implications for understanding how promoters are located within physiological settings.
Figures

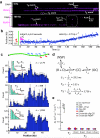
 ), and magenta bars highlight the first 3–9 seconds of the reaction trajectory. (c) Binding distributions of kinetically distinct intermediates, and corresponding lifetime measurements (insets; also see Supplementary Fig. 2 & Supplementary Fig. 2); a schematic showing the relative promoter location is included. Error bars indicate 70% confidence intervals obtained through bootstrap analysis.
(d) Kinetic scheme reflecting observed intermediates. NSP, CC, and OC, refer to nonspecifically bound, closed complex, and open complex, respectively; note that CC could also represent another intermediate preceding the open complex. Kinetic parameters are not segregated for individual promoters, rather they are considered collectively, and therefore reported values should be considered an average of all λ promoters. (e) Upper bound of observed diffusion coefficients for promoter–bound RNAP, compared to immobilized dig–QDs and other proteins known to undergo 1D–diffusion (Supplementary Fig. 7–8 & Supplementary Table 4).,, Diffusion coefficients are gamma distributed, therefore we report the magnitude of the square root of the variance as error bars (n ≥ 50 for all data sets).
), and magenta bars highlight the first 3–9 seconds of the reaction trajectory. (c) Binding distributions of kinetically distinct intermediates, and corresponding lifetime measurements (insets; also see Supplementary Fig. 2 & Supplementary Fig. 2); a schematic showing the relative promoter location is included. Error bars indicate 70% confidence intervals obtained through bootstrap analysis.
(d) Kinetic scheme reflecting observed intermediates. NSP, CC, and OC, refer to nonspecifically bound, closed complex, and open complex, respectively; note that CC could also represent another intermediate preceding the open complex. Kinetic parameters are not segregated for individual promoters, rather they are considered collectively, and therefore reported values should be considered an average of all λ promoters. (e) Upper bound of observed diffusion coefficients for promoter–bound RNAP, compared to immobilized dig–QDs and other proteins known to undergo 1D–diffusion (Supplementary Fig. 7–8 & Supplementary Table 4).,, Diffusion coefficients are gamma distributed, therefore we report the magnitude of the square root of the variance as error bars (n ≥ 50 for all data sets).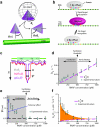
 ) correspond to proteins that fail to bind the operator, magenta data points (
) correspond to proteins that fail to bind the operator, magenta data points ( ) are proteins that bind the operator after undergoing FD, and green data points (
) are proteins that bind the operator after undergoing FD, and green data points ( ) correspond 3D binding to the operator. All green data points within each column overlap at zero, but their fractional contribution to operator binding is shown as green bars in the inset of panel (e). These experiments were all conducted in buffer containing 10 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 1 mM DTT, and 1 mg ml−1 BSA.
) correspond 3D binding to the operator. All green data points within each column overlap at zero, but their fractional contribution to operator binding is shown as green bars in the inset of panel (e). These experiments were all conducted in buffer containing 10 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 1 mM DTT, and 1 mg ml−1 BSA.
Comment in
-
Looking for a promoter in 3D.Nat Struct Mol Biol. 2013 Feb;20(2):141-2. doi: 10.1038/nsmb.2498. Nat Struct Mol Biol. 2013. PMID: 23381630 No abstract available.
References
-
- von Hippel P, Berg O. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–8. - PubMed
-
- Gorman J, Greene EC. Visualizing one–dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–74. - PubMed
-
- Browning D, Busby S. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

