Chromatin marks identify critical cell types for fine mapping complex trait variants
- PMID: 23263488
- PMCID: PMC3826950
- DOI: 10.1038/ng.2504
Chromatin marks identify critical cell types for fine mapping complex trait variants
Abstract
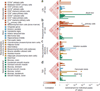
If trait-associated variants alter regulatory regions, then they should fall within chromatin marks in relevant cell types. However, it is unclear which of the many marks are most useful in defining cell types associated with disease and fine mapping variants. We hypothesized that informative marks are phenotypically cell type specific; that is, SNPs associated with the same trait likely overlap marks in the same cell type. We examined 15 chromatin marks and found that those highlighting active gene regulation were phenotypically cell type specific. Trimethylation of histone H3 at lysine 4 (H3K4me3) was the most phenotypically cell type specific (P < 1 × 10(-6)), driven by colocalization of variants and marks rather than gene proximity (P < 0.001). H3K4me3 peaks overlapped with 37 SNPs for plasma low-density lipoprotein concentration in the liver (P < 7 × 10(-5)), 31 SNPs for rheumatoid arthritis within CD4(+) regulatory T cells (P = 1 × 10(-4)), 67 SNPs for type 2 diabetes in pancreatic islet cells (P = 0.003) and the liver (P = 0.003), and 14 SNPs for neuropsychiatric disease in neuronal tissues (P = 0.007). We show how cell type-specific H3K4me3 peaks can inform the fine mapping of associated SNPs to identify causal variation.
Figures



References
-
- Fraser HB, Xie X. Common polymorphic transcript variation in human disease. Genome Res. 2009;19:567–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

