Single-molecule analysis of gene expression using two-color RNA labeling in live yeast
- PMID: 23263691
- PMCID: PMC3899799
- DOI: 10.1038/nmeth.2305
Single-molecule analysis of gene expression using two-color RNA labeling in live yeast
Abstract
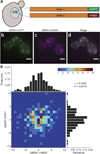
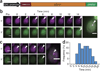
Live-cell imaging of mRNA yields important insights into gene expression, but it has generally been limited to the labeling of one RNA species and has never been used to count single mRNAs over time in yeast. We demonstrate a two-color imaging system with single-molecule resolution using MS2 and PP7 RNA labeling. We use this methodology to measure intrinsic noise in mRNA levels and RNA polymerase II kinetics at a single gene.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

