Phylogenetic distribution of potential cellulases in bacteria
- PMID: 23263967
- PMCID: PMC3591946
- DOI: 10.1128/AEM.03305-12
Phylogenetic distribution of potential cellulases in bacteria
Abstract
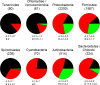
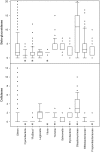
Many microorganisms contain cellulases that are important for plant cell wall degradation and overall soil ecosystem functioning. At present, we have extensive biochemical knowledge of cellulases but little is known about the phylogenetic distribution of these enzymes. To address this, we analyzed the distribution of 21,985 genes encoding proteins related to cellulose utilization in 5,123 sequenced bacterial genomes. First, we identified the distribution of glycoside hydrolases involved in cellulose utilization and synthesis at different taxonomic levels, from the phylum to the strain. Cellulose degradation/utilization capabilities appeared in nearly all major groups and resulted in strains displaying various enzyme gene combinations. Potential cellulose degraders, having both cellulases and β-glucosidases, constituted 24% of all genomes whereas potential opportunistic strains, having β-glucosidases only, accounted for 56%. Finally, 20% of the bacteria have no relevant enzymes and do not rely on cellulose utilization. The latter group was primarily connected to specific bacterial lifestyles like autotrophy and parasitism. Cellulose degraders, as well as opportunists, have multiple enzymes with similar functions. However, the potential degraders systematically harbor about twice more β-glucosidases than their potential opportunistic relatives. Although scattered, the distribution of functional types, in bacterial lineages, is not random but mostly follows a Brownian motion evolution model. Degraders form clusters of relatives at the species level, whereas opportunists are clustered at the genus level. This information can form a mechanistic basis for the linking of changes in microbial community composition to soil ecosystem processes.
Figures



References
-
- Wilson DB. 2011. Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14:259–263 - PubMed
-
- Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front. Microbiol. 2:94 doi:10.3389/fmicb.2011.00094 - DOI - PMC - PubMed
-
- Allison SD. 2005. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett. 8:626–635
-
- Romaní AM, Fischer H, Mille-Lindblom C, Tranvik LJ. 2006. Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology 87:2559–2569 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

