Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos
- PMID: 23263990
- PMCID: PMC3623074
- DOI: 10.1128/MCB.01014-12
Transient JMJD2B-mediated reduction of H3K9me3 levels improves reprogramming of embryonic stem cells into cloned embryos
Abstract
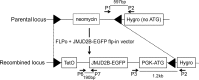
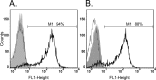
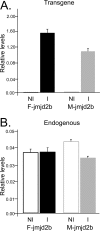
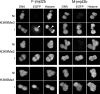
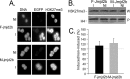
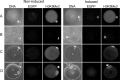
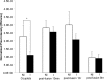
Correct reprogramming of epigenetic marks in the donor nuclei is crucial for successful cloning by nuclear transfer. Specific epigenetic modifications, such as repressive histone lysine methylation marks, are known to be very stable and difficult to reprogram. The discovery of histone lysine demethylases has opened up opportunities to study the effects of removing repressive histone lysine methylation marks in donor cells prior to nuclear transfer. In this study, we generated mouse embryonic stem (ES) cells for the inducible expression of JMJD2B (also known as KDM4B), a demethylase that primarily removes the histone-3 lysine-9 trimethylation (H3K9me3) mark. Induction of jmjd2b in the ES cells decreased total levels of H3K9me3 by 63%. When these cells were used for nuclear transfer, H3K9me3 levels were normalized within minutes following fusion with an enucleated oocyte. This transient reduction of H3K9me3 levels improved in vitro development into cloned embryos by 30%.
Figures

References
-
- Armstrong L, Lako M, Dean W, Stojkovic M. 2006. Epigenetic modification is central to genome reprogramming in somatic cell nuclear transfer. Stem Cells 24: 805– 814 - PubMed
-
- Fujita N, Wade PA. 2004. Nuclear transfer: epigenetics pays a visit. Nat. Cell Biol. 6: 920– 922 - PubMed
-
- Morgan HD, Santos F, Green K, Dean W, Reik W. 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14(Spec No. 1):R47–R58 - PubMed
-
- Rideout WM, III, Eggan K, Jaenisch R. 2001. Nuclear cloning and epigenetic reprogramming of the genome. Science 293: 1093– 1098 - PubMed
-
- Santos F, Dean W. 2004. Epigenetic reprogramming during early development in mammals. Reproduction 127: 643– 651 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
