PDGFRα signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways
- PMID: 23264740
- PMCID: PMC4481637
- DOI: 10.1242/jcs.116426
PDGFRα signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways
Abstract
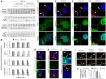
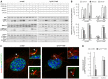
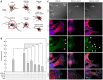
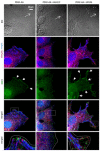
In fibroblasts, platelet-derived growth factor receptor alpha (PDGFRα) is upregulated during growth arrest and compartmentalized to the primary cilium. PDGF-AA mediated activation of the dimerized ciliary receptor produces a phosphorylation cascade through the PI3K-AKT and MEK1/2-ERK1/2 pathways leading to the activation of the Na(+)/H(+) exchanger, NHE1, cytoplasmic alkalinization and actin nucleation at the lamellipodium that supports directional cell migration. We here show that AKT and MEK1/2-ERK1/2-p90(RSK) inhibition reduced PDGF-AA-induced cell migration by distinct mechanisms: AKT inhibition reduced NHE1 activity by blocking the translocation of NHE1 to the cell membrane. MEK1/2 inhibition did not affect NHE1 activity but influenced NHE1 localization, causing NHE1 to localize discontinuously in patches along the plasma membrane, rather than preferentially at the lamellipodium. We also provide direct evidence of NHE1 translocation through the cytoplasm to the leading edge. In conclusion, signals initiated at the primary cilium through the PDGFRαα cascade reorganize the cytoskeleton to regulate cell migration differentially through the AKT and the MEK1/2-ERK1/2-p90(RSK) pathways. The AKT pathway is necessary for initiation of NHE1 translocation, presumably in vesicles, to the leading edge and for its activation. In contrast, the MEK1/2-ERK1/2-p90(RSK) pathway controls the spatial organization of NHE1 translocation and incorporation, and therefore specifies the direction of the leading edge formation.
Figures



References
-
- Boedtkjer E., Bunch L., Pedersen S. F. (2012). Physiology, pharmacology and pathophysiology of the pH regulatory transport proteins NHE1 and NBCn1: similarities, differences, and implications for cancer therapy. Curr. Pharm. Des. 18, 1345–1371. - PubMed
-
- Boron W. F. (2004). Regulation of intracellular pH. Adv. Physiol. Educ. 28, 160–179. - PubMed
-
- Boyarsky G., Ganz M. B., Sterzel R. B., Boron W. F. (1988). pH regulation in single glomerular mesangial cells. II. Na+-dependent and -independent Cl(-)-HCO3- exchangers. Am. J. Physiol. 255, C857–C869. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

