Identification of genome-wide copy number variations among diverse pig breeds by array CGH
- PMID: 23265576
- PMCID: PMC3573951
- DOI: 10.1186/1471-2164-13-725
Identification of genome-wide copy number variations among diverse pig breeds by array CGH
Abstract
Background: Recent studies have shown that copy number variation (CNV) in mammalian genomes contributes to phenotypic diversity, including health and disease status. In domestic pigs, CNV has been catalogued by several reports, but the extent of CNV and the phenotypic effects are far from clear. The goal of this study was to identify CNV regions (CNVRs) in pigs based on array comparative genome hybridization (aCGH).
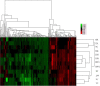
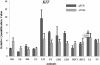
Results: Here a custom-made tiling oligo-nucleotide array was used with a median probe spacing of 2506 bp for screening 12 pigs including 3 Chinese native pigs (one Chinese Erhualian, one Tongcheng and one Yangxin pig), 5 European pigs (one Large White, one Pietrain, one White Duroc and two Landrace pigs), 2 synthetic pigs (Chinese new line DIV pigs) and 2 crossbred pigs (Landrace × DIV pigs) with a Duroc pig as the reference. Two hundred and fifty-nine CNVRs across chromosomes 1-18 and X were identified, with an average size of 65.07 kb and a median size of 98.74 kb, covering 16.85 Mb or 0.74% of the whole genome. Concerning copy number status, 93 (35.91%) CNVRs were called as gains, 140 (54.05%) were called as losses and the remaining 26 (10.04%) were called as both gains and losses. Of all detected CNVRs, 171 (66.02%) and 34 (13.13%) CNVRs directly overlapped with Sus scrofa duplicated sequences and pig QTLs, respectively. The CNVRs encompassed 372 full length Ensembl transcripts. Two CNVRs identified by aCGH were validated using real-time quantitative PCR (qPCR).
Conclusions: Using 720 K array CGH (aCGH) we described a map of porcine CNVs which facilitated the identification of structural variations for important phenotypes and the assessment of the genetic diversity of pigs.
Figures

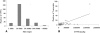
References
-
- Amills M, Clop A, Ramı′rez O, Pe′rez-Enciso M. Encyclopedia of Life Sciences (ELS) John Wiley & Sons, Ltd, Chichester; 2010. Origin and genetic diversity of pig breeds.
-
- Haley CS, Agaro E, Ellis M. Genetic components of growth and ultrasonic fat depth traits in Meishan and Large White pigs and their reciprocal crosses. Anim Prod. 1992;54:105–115. doi: 10.1017/S0003356100020626. - DOI
-
- Haley CS, Lee GJ, Ritchie M. Comparative reproductive-performance in Meishan and Large White pigs and threir crosses. Anim Sci. 1995;60:259–267. doi: 10.1017/S1357729800008420. - DOI
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

