The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks
- PMID: 23266955
- PMCID: PMC3529529
- DOI: 10.1083/jcb.201205059
The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks
Abstract
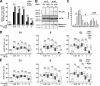
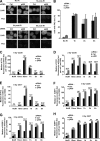
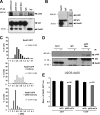
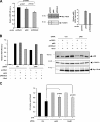
DNA damage signaling and repair take place in a chromatin context. Consequently, chromatin-modifying enzymes, including adenosine triphosphate-dependent chromatin remodeling enzymes, play an important role in the management of DNA double-strand breaks (DSBs). Here, we show that the p400 ATPase is required for DNA repair by homologous recombination (HR). Indeed, although p400 is not required for DNA damage signaling, DNA DSB repair is defective in the absence of p400. We demonstrate that p400 is important for HR-dependent processes, such as recruitment of Rad51 to DSB (a key component of HR), homology-directed repair, and survival after DNA damage. Strikingly, p400 and Rad51 are present in the same complex and both favor chromatin remodeling around DSBs. Altogether, our data provide a direct molecular link between Rad51 and a chromatin remodeling enzyme involved in chromatin decompaction around DNA DSBs.
Figures





References
-
- Brunton H., Goodarzi A.A., Noon A.T., Shrikhande A., Hansen R.S., Jeggo P.A., Shibata A. 2011. Analysis of human syndromes with disordered chromatin reveals the impact of heterochromatin on the efficacy of ATM-dependent G2/M checkpoint arrest. Mol. Cell. Biol. 31:4022–4035 10.1128/MCB.05289-11 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

