Modular assembly of yeast cytochrome oxidase
- PMID: 23266989
- PMCID: PMC3571867
- DOI: 10.1091/mbc.E12-10-0749
Modular assembly of yeast cytochrome oxidase
Abstract
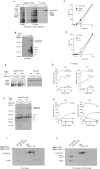
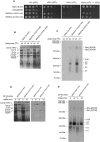
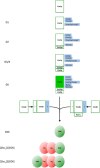
Previous studies of yeast cytochrome oxidase (COX) biogenesis identified Cox1p, one of the three mitochondrially encoded core subunits, in two high-molecular weight complexes combined with regulatory/assembly factors essential for expression of this subunit. In the present study we use pulse-chase labeling experiments in conjunction with isolated mitochondria to identify new Cox1p intermediates and place them in an ordered pathway. Our results indicate that before its assimilation into COX, Cox1p transitions through five intermediates that are differentiated by their compositions of accessory factors and of two of the eight imported subunits. We propose a model of COX biogenesis in which Cox1p and the two other mitochondrial gene products, Cox2p and Cox3p, constitute independent assembly modules, each with its own complement of subunits. Unlike their bacterial counterparts, which are composed only of the individual core subunits, the final sequence in which the mitochondrial modules associate to form the holoenzyme may have been conserved during evolution.
Figures




References
-
- Carr HS, Winge DR. Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res. 2003;36:309–316. - PubMed
-
- Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA. Assembly-controlled regulation of chloroplast gene translation. Biochem Soc Trans. 2001;29:421–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

