A novel non-radioactive primase-pyrophosphatase activity assay and its application to the discovery of inhibitors of Mycobacterium tuberculosis primase DnaG
- PMID: 23267008
- PMCID: PMC3575809
- DOI: 10.1093/nar/gks1292
A novel non-radioactive primase-pyrophosphatase activity assay and its application to the discovery of inhibitors of Mycobacterium tuberculosis primase DnaG
Abstract
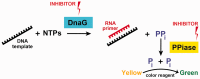
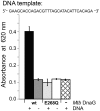
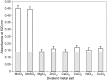
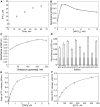
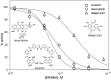
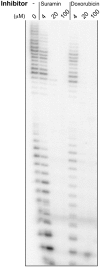
Bacterial DNA primase DnaG synthesizes RNA primers required for chromosomal DNA replication. Biochemical assays measuring primase activity have been limited to monitoring formation of radioactively labelled primers because of the intrinsically low catalytic efficiency of DnaG. Furthermore, DnaG is prone to aggregation and proteolytic degradation. These factors have impeded discovery of DnaG inhibitors by high-throughput screening (HTS). In this study, we expressed and purified the previously uncharacterized primase DnaG from Mycobacterium tuberculosis (Mtb DnaG). By coupling the activity of Mtb DnaG to that of another essential enzyme, inorganic pyrophosphatase from M. tuberculosis (Mtb PPiase), we developed the first non-radioactive primase-pyrophosphatase assay. An extensive optimization of the assay enabled its efficient use in HTS (Z' = 0.7 in the 384-well format). HTS of 2560 small molecules to search for inhibitory compounds yielded several hits, including suramin, doxorubicin and ellagic acid. We demonstrate that these three compounds inhibit Mtb DnaG. Both suramin and doxorubicin are potent (low-µM) DNA- and nucleotide triphosphate-competitive priming inhibitors that interact with more than one site on Mtb DnaG. This novel assay should be applicable to other primases and inefficient DNA/RNA polymerases, facilitating their characterization and inhibitor discovery.
Figures



Similar articles
-
Antimycobacterial activity of DNA intercalator inhibitors of Mycobacterium tuberculosis primase DnaG.J Antibiot (Tokyo). 2015 Mar;68(3):153-7. doi: 10.1038/ja.2014.131. Epub 2014 Sep 24. J Antibiot (Tokyo). 2015. PMID: 25248725 Free PMC article.
-
Discovery of inhibitors of Bacillus anthracis primase DnaG.Biochemistry. 2013 Oct 1;52(39):6905-10. doi: 10.1021/bi4011286. Epub 2013 Sep 19. Biochemistry. 2013. PMID: 24004110
-
Evaluation of DNA primase DnaG as a potential target for antibiotics.Antimicrob Agents Chemother. 2014;58(3):1699-706. doi: 10.1128/AAC.01721-13. Epub 2013 Dec 30. Antimicrob Agents Chemother. 2014. PMID: 24379196 Free PMC article.
-
DnaG Primase-A Target for the Development of Novel Antibacterial Agents.Antibiotics (Basel). 2018 Aug 13;7(3):72. doi: 10.3390/antibiotics7030072. Antibiotics (Basel). 2018. PMID: 30104489 Free PMC article. Review.
-
Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions.Nucleic Acids Res. 2006;34(15):4082-8. doi: 10.1093/nar/gkl363. Epub 2006 Aug 25. Nucleic Acids Res. 2006. PMID: 16935873 Free PMC article. Review.
Cited by
-
Modulation of RNA primer formation by Mn(II)-substituted T7 DNA primase.Sci Rep. 2017 Jul 19;7(1):5797. doi: 10.1038/s41598-017-05534-3. Sci Rep. 2017. PMID: 28724886 Free PMC article.
-
Antimycobacterial activity of DNA intercalator inhibitors of Mycobacterium tuberculosis primase DnaG.J Antibiot (Tokyo). 2015 Mar;68(3):153-7. doi: 10.1038/ja.2014.131. Epub 2014 Sep 24. J Antibiot (Tokyo). 2015. PMID: 25248725 Free PMC article.
-
A Colorimetric Assay to Identify and Characterize Bacterial Primase Inhibitors.Methods Mol Biol. 2023;2601:283-301. doi: 10.1007/978-1-0716-2855-3_15. Methods Mol Biol. 2023. PMID: 36445590
-
CRISPR-Associated Primase-Polymerases are implicated in prokaryotic CRISPR-Cas adaptation.Nat Commun. 2021 Jun 17;12(1):3690. doi: 10.1038/s41467-021-23535-9. Nat Commun. 2021. PMID: 34140468 Free PMC article.
-
2',6'-Dihalostyrylanilines, pyridines, and pyrimidines for the inhibition of the catalytic subunit of methionine S-adenosyltransferase-2.J Med Chem. 2014 Jul 24;57(14):6083-91. doi: 10.1021/jm5004864. Epub 2014 Jul 9. J Med Chem. 2014. PMID: 24950374 Free PMC article.
References
-
- Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. - PubMed
-
- Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. - PubMed
-
- Augustin MA, Huber R, Kaiser JT. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat. Struct. Biol. 2001;8:57–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

