Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure?
- PMID: 23267412
- PMCID: PMC3527312
- DOI: 10.4161/bioa.22592
Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure?
Abstract
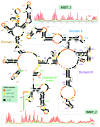
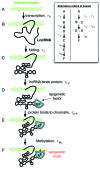
Long noncoding RNAs (lncRNAs) play a key role in many important areas of epigenetics, stem cell biology, cancer, signaling and brain function. This emerging class of RNAs constitutes a large fraction of the transcriptome, with thousands of new lncRNAs reported each year. The molecular mechanisms of these RNAs are not well understood. Currently, very little structural data exist. We review the available lncRNA sequence and secondary structure data. Since almost no tertiary information is available for lncRNAs, we review crystallographic structures for other RNA systems and discuss the possibilities for lncRNAs in the context of existing constraints.
Keywords: HOTAIR; MALAT; RNA; RNA structure; cancer; epigenetics; hormone receptor; lincRNA; lncRNA; long noncoding RNA; non-coding; secondary structure; structural biology.
Figures
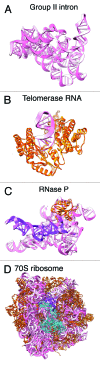

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
