A survey of host range genes in poxvirus genomes
- PMID: 23268114
- PMCID: PMC4080715
- DOI: 10.1016/j.meegid.2012.12.002
A survey of host range genes in poxvirus genomes
Abstract
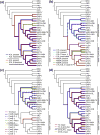
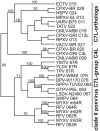
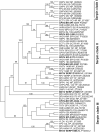
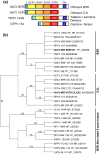
Poxviruses are widespread pathogens, which display extremely different host ranges. Whereas some poxviruses, including variola virus, display narrow host ranges, others such as cowpox viruses naturally infect a wide range of mammals. The molecular basis for differences in host range are poorly understood but apparently depend on the successful manipulation of the host antiviral response. Some poxvirus genes have been shown to confer host tropism in experimental settings and are thus called host range factors. Identified host range genes include vaccinia virus K1L, K3L, E3L, B5R, C7L and SPI-1, cowpox virus CP77/CHOhr, ectromelia virus p28 and 022, and myxoma virus T2, T4, T5, 11L, 13L, 062R and 063R. These genes encode for ankyrin repeat-containing proteins, tumor necrosis factor receptor II homologs, apoptosis inhibitor T4-related proteins, Bcl-2-related proteins, pyrin domain-containing proteins, cellular serine protease inhibitors (serpins), short complement-like repeats containing proteins, KilA-N/RING domain-containing proteins, as well as inhibitors of the double-stranded RNA-activated protein kinase PKR. We conducted a systematic survey for the presence of known host range genes and closely related family members in poxvirus genomes, classified them into subgroups based on their phylogenetic relationship and correlated their presence with the poxvirus phylogeny. Common themes in the evolution of poxvirus host range genes are lineage-specific duplications and multiple independent inactivation events. Our analyses yield new insights into the evolution of poxvirus host range genes. Implications of our findings for poxvirus host range and virulence are discussed.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Poxviruses and the evolution of host range and virulence.Infect Genet Evol. 2014 Jan;21:15-40. doi: 10.1016/j.meegid.2013.10.014. Epub 2013 Oct 24. Infect Genet Evol. 2014. PMID: 24161410 Free PMC article. Review.
-
Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L.Virology. 2008 Mar 15;372(2):372-83. doi: 10.1016/j.virol.2007.10.023. Epub 2007 Dec 3. Virology. 2008. PMID: 18054061 Free PMC article.
-
Sequence and phylogenetic analysis of host-range (E3L, K3L, and C7L) and structural protein (B5R) genes of buffalopox virus isolates from buffalo, cattle, and human in India.Virus Genes. 2012 Dec;45(3):488-98. doi: 10.1007/s11262-012-0788-8. Epub 2012 Aug 8. Virus Genes. 2012. PMID: 22872567
-
Poxvirus genome evolution by gene gain and loss.Mol Phylogenet Evol. 2005 Apr;35(1):186-95. doi: 10.1016/j.ympev.2004.12.008. Epub 2005 Jan 21. Mol Phylogenet Evol. 2005. PMID: 15737590
-
Poxvirus Host Range Genes and Virus-Host Spectrum: A Critical Review.Viruses. 2017 Nov 7;9(11):331. doi: 10.3390/v9110331. Viruses. 2017. PMID: 29112165 Free PMC article. Review.
Cited by
-
Novel Class of Viral Ankyrin Proteins Targeting the Host E3 Ubiquitin Ligase Cullin-2.J Virol. 2018 Nov 12;92(23):e01374-18. doi: 10.1128/JVI.01374-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30258003 Free PMC article.
-
Cowpox Viruses: A Zoo Full of Viral Diversity and Lurking Threats.Biomolecules. 2023 Feb 8;13(2):325. doi: 10.3390/biom13020325. Biomolecules. 2023. PMID: 36830694 Free PMC article. Review.
-
Monkeypox: Immune response, vaccination and preventive efforts.Narra J. 2022 Dec;2(3):e90. doi: 10.52225/narra.v2i3.90. Epub 2022 Oct 22. Narra J. 2022. PMID: 38449905 Free PMC article. Review.
-
Whole genome sequencing of recombinant viruses obtained from co-infection and superinfection of Vero cells with modified vaccinia virus ankara vectored influenza vaccine and a naturally occurring cowpox virus.Front Immunol. 2024 Apr 3;15:1277447. doi: 10.3389/fimmu.2024.1277447. eCollection 2024. Front Immunol. 2024. PMID: 38633245 Free PMC article.
-
Deletion of the Vaccinia Virus B1 Kinase Reveals Essential Functions of This Enzyme Complemented Partly by the Homologous Cellular Kinase VRK2.J Virol. 2017 Jul 12;91(15):e00635-17. doi: 10.1128/JVI.00635-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515294 Free PMC article.
References
-
- Ali AN, Turner PC, Brooks MA, Moyer RW. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:305–314. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

