STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis
- PMID: 23268445
- PMCID: PMC3561960
- DOI: 10.1093/nar/gks1309
STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis
Abstract
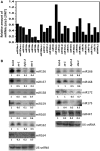
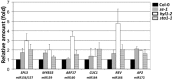
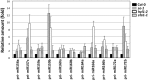
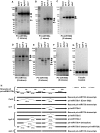
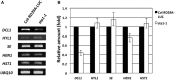
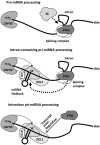
MicroRNAs (miRNAs) are small regulatory RNAs that have important regulatory roles in numerous developmental and metabolic processes in most eukaryotes. In Arabidopsis, DICER-LIKE1 (DCL1), HYPONASTIC LEAVES 1, SERRATE, HUA ENHANCER1 and HASTY are involved in processing of primary miRNAs (pri-miRNAs) to yield precursor miRNAs (pre-miRNAs) and eventually miRNAs. In addition to these components, mRNA cap-binding proteins, CBP80/ABA HYPERSENSITIVE1 and CBP20, also participate in miRNA biogenesis. Here, we show that STABILIZED1 (STA1), an Arabidopsis pre-mRNA processing factor 6 homolog, is also involved in the biogenesis of miRNAs. Similar to other miRNA biogenesis-defective mutants, sta1-1 accumulated significantly lower levels of mature miRNAs and concurrently higher levels of pri-miRNAs than wild type. The dramatic reductions of mature miRNAs were associated with the accumulation of their target gene transcripts and developmental defects. Furthermore, sta1-1 impaired splicing of intron containing pri-miRNAs and decreased transcript levels of DCL1. These results suggest that STA1 is involved in miRNA biogenesis directly by functioning in pri-miRNA splicing and indirectly by modulating the DCL1 transcript level.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

