A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent
- PMID: 23268449
- PMCID: PMC3561988
- DOI: 10.1093/nar/gks1282
A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent
Abstract
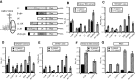
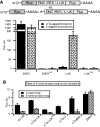
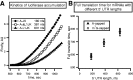
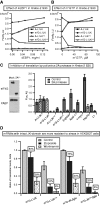
Resistance of translation of some eukaryotic messenger RNAs (mRNAs) to inactivation of the cap-binding factor eIF4E under unfavorable conditions is well documented. To date, it is the mechanism of internal ribosome entry that is predominantly thought to underlay this stress tolerance. However, many cellular mRNAs that had been considered to contain internal ribosome entry sites (IRESs) failed to pass stringent control tests for internal initiation, thus raising the question of how they are translated under stress conditions. Here, we show that inserting an eIF4G-binding element from a virus IRES into 5'-UTRs of strongly cap-dependent mRNAs dramatically reduces their requirement for the 5'-terminal m(7)G-cap, though such cap-independent translation remains dependent on a vacant 5'-terminus of these mRNAs. Importantly, direct binding of eIF4G to the 5'-UTR of mRNA makes its translation resistant to eIF4F inactivation both in vitro and in vivo. These data may substantiate a new paradigm of translational control under stress to complement IRES-driven mechanism of translation.
Figures

References
-
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

