Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions
- PMID: 23269503
- PMCID: PMC3596480
- DOI: 10.1007/s11095-012-0935-x
Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions
Abstract
Purpose: Membrane transporters mediate many biological effects of chemicals and play a major role in pharmacokinetics and drug resistance. The selection of viable drug candidates among biologically active compounds requires the assessment of their transporter interaction profiles.
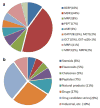
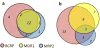
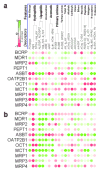
Methods: Using public sources, we have assembled and curated the largest, to our knowledge, human intestinal transporter database (>5,000 interaction entries for >3,700 molecules). This data was used to develop thoroughly validated classification Quantitative Structure-Activity Relationship (QSAR) models of transport and/or inhibition of several major transporters including MDR1, BCRP, MRP1-4, PEPT1, ASBT, OATP2B1, OCT1, and MCT1.
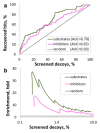
Results: QSAR models have been developed with advanced machine learning techniques such as Support Vector Machines, Random Forest, and k Nearest Neighbors using Dragon and MOE chemical descriptors. These models afforded high external prediction accuracies of 71-100% estimated by 5-fold external validation, and showed hit retrieval rates with up to 20-fold enrichment in the virtual screening of DrugBank compounds.
Conclusions: The compendium of predictive QSAR models developed in this study can be used for virtual profiling of drug candidates and/or environmental agents with the optimal transporter profiles.
Figures

References
-
- Marquez B, Van Bambeke F. ABC multidrug transporters: target for modulation of drug pharmacokinetics and drug-drug interactions. Curr Drug Targets. 2011;12:600–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

