Biological and phylogenetic characteristics of yellow fever virus lineages from West Africa
- PMID: 23269797
- PMCID: PMC3571399
- DOI: 10.1128/JVI.01116-12
Biological and phylogenetic characteristics of yellow fever virus lineages from West Africa
Abstract
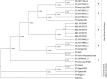
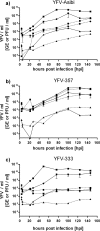
The yellow fever virus (YFV), the first proven human-pathogenic virus, although isolated in 1927, is still a major public health problem, especially in West Africa where it causes outbreaks every year. Nevertheless, little is known about its genetic diversity and evolutionary dynamics, mainly due to a limited number of genomic sequences from wild virus isolates. In this study, we analyzed the phylogenetic relationships of 24 full-length genomes from YFV strains isolated between 1973 and 2005 in a sylvatic context of West Africa, including 14 isolates that had previously not been sequenced. By this, we confirmed genetic variability within one genotype by the identification of various YF lineages circulating in West Africa. Further analyses of the biological properties of these lineages revealed differential growth behavior in human liver and insect cells, correlating with the source of isolation and suggesting host adaptation. For one lineage, repeatedly isolated in a context of vertical transmission, specific characteristics in the growth behavior and unique mutations of the viral genome were observed and deserve further investigation to gain insight into mechanisms involved in YFV emergence and maintenance in nature.
Figures





Similar articles
-
Genome analysis and phylogenetic relationships between east, central and west African isolates of Yellow fever virus.J Gen Virol. 2006 Apr;87(Pt 4):895-907. doi: 10.1099/vir.0.81236-0. J Gen Virol. 2006. PMID: 16528039
-
Phylogeographic reconstruction of African yellow fever virus isolates indicates recent simultaneous dispersal into east and west Africa.PLoS Negl Trop Dis. 2013;7(3):e1910. doi: 10.1371/journal.pntd.0001910. Epub 2013 Mar 14. PLoS Negl Trop Dis. 2013. PMID: 23516640 Free PMC article. Review.
-
Molecular epidemiology of yellow fever in Bolivia from 1999 to 2008.Vector Borne Zoonotic Dis. 2011 Mar;11(3):277-84. doi: 10.1089/vbz.2010.0017. Epub 2010 Oct 6. Vector Borne Zoonotic Dis. 2011. PMID: 20925524 Free PMC article.
-
Nucleotide sequence variation of the envelope protein gene identifies two distinct genotypes of yellow fever virus.J Virol. 1995 Sep;69(9):5773-80. doi: 10.1128/JVI.69.9.5773-5780.1995. J Virol. 1995. PMID: 7637022 Free PMC article.
-
The enigma of yellow fever in East Africa.Rev Med Virol. 2008 Sep-Oct;18(5):331-46. doi: 10.1002/rmv.584. Rev Med Virol. 2008. PMID: 18615782 Review.
Cited by
-
Molecular Characterization of Circulating Yellow Fever Viruses from Outbreak in Ghana, 2021-2022.Emerg Infect Dis. 2023 Sep;29(9):1818-1826. doi: 10.3201/eid2909.221671. Emerg Infect Dis. 2023. PMID: 37610174 Free PMC article.
-
The Brazilian Zika virus strain causes birth defects in experimental models.Nature. 2016 Jun 9;534(7606):267-71. doi: 10.1038/nature18296. Epub 2016 May 11. Nature. 2016. PMID: 27279226 Free PMC article.
-
Genomic sequence of yellow fever virus from a Dutch traveller returning from the Gambia-Senegal region, the Netherlands, November 2018.Euro Surveill. 2019 Jan;24(4):1800684. doi: 10.2807/1560-7917.ES.2019.24.4.1800684. Euro Surveill. 2019. PMID: 30696531 Free PMC article.
-
Evaluating vector competence for Yellow fever in the Caribbean.Nat Commun. 2024 Feb 9;15(1):1236. doi: 10.1038/s41467-024-45116-2. Nat Commun. 2024. PMID: 38336944 Free PMC article.
-
Phylogenomic analysis unravels evolution of yellow fever virus within hosts.PLoS Negl Trop Dis. 2018 Sep 6;12(9):e0006738. doi: 10.1371/journal.pntd.0006738. eCollection 2018 Sep. PLoS Negl Trop Dis. 2018. PMID: 30188905 Free PMC article.
References
-
- Monath TP. 2008. Treatment of yellow fever. Antiviral Res. 78:116–124 - PubMed
-
- Monath TP. 2001. Yellow fever: an update. Lancet Infect. Dis. 1:11–20 - PubMed
-
- Mutebi JP, Barrett AD. 2002. The epidemiology of yellow fever in Africa. Microbes Infect. 4:1459–1468 - PubMed
-
- Barrett AD, Monath TP. 2003. Epidemiology and ecology of yellow fever virus. Adv. Virus Res. 61:291–315 - PubMed
-
- Robertson SE, Hull BP, Tomori O, Bele O, LeDuc JW, Esteves K. 1996. Yellow fever: a decade of reemergence. JAMA 276:1157–1162 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

