A versatile mass spectrometry-based method to both identify kinase client-relationships and characterize signaling network topology
- PMID: 23270405
- PMCID: PMC3888875
- DOI: 10.1021/pr3009995
A versatile mass spectrometry-based method to both identify kinase client-relationships and characterize signaling network topology
Abstract
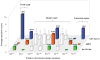
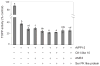
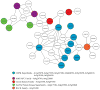
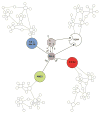
While more than a thousand protein kinases (PK) have been identified in the Arabidopsis thaliana genome, relatively little progress has been made toward identifying their individual client proteins. Herein we describe the use of a mass spectrometry-based in vitro phosphorylation strategy, termed Kinase Client assay (KiC assay), to study a targeted-aspect of signaling. A synthetic peptide library comprising 377 in vivo phosphorylation sequences from developing seed was screened using 71 recombinant A. thaliana PK. Among the initial results, we identified 23 proteins as putative clients of 17 PK. In one instance protein phosphatase inhibitor-2 (AtPPI-2) was phosphorylated at multiple-sites by three distinct PK, casein kinase1-like 10, AME3, and a Ser PK-like protein. To confirm this result, full-length recombinant AtPPI-2 was reconstituted with each of these PK. The results confirmed multiple distinct phosphorylation sites within this protein. Biochemical analyses indicate that AtPPI-2 inhibits type 1 protein phosphatase (TOPP) activity, and that the phosphorylated forms of AtPPI-2 are more potent inhibitors. Structural modeling revealed that phosphorylation of AtPPI-2 induces conformational changes that modulate TOPP binding.
Figures



References
-
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. - PMC - PubMed
-
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, Gendron JM, Jonassen EM, Lillo C, DeLong A, Burlingame AL, Sun Y, Wang ZY. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–131. - PMC - PubMed
-
- Cohen PTW. Protein phosphatase 1: Targeted in many directions. J Cell Sci. 2002;115:241–256. - PubMed
-
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008;55:104–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

