Enrichment of meiotic recombination hotspot sequences by avidin capture technology
- PMID: 23270922
- PMCID: PMC3563735
- DOI: 10.1016/j.gene.2012.12.042
Enrichment of meiotic recombination hotspot sequences by avidin capture technology
Abstract
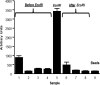
About 40% of the hotspots for meiotic recombination contain the degenerate consensus sequence 5'-CCNCCNTNNCCNC-3'. Here we present a novel protocol for enriching hotspot sequences from digested genomic DNA by using biotinylated oligonucleotides and streptavidin-coated magnetic beads. The captured hotspots can be released by simple digestion with restriction enzymes for subsequent characterization by second generation sequencing or PCR. The capture protocol specifically enriches hotspot sequences, judged by using fluorophore-conjugated synthetic oligonucleotides and synthetic double-stranded oligonucleotides in combination with PCR. The capture protocol enriches single-stranded DNA, denatured double-stranded DNA, and large fragments (>3000 bp) of digested plasmid DNA with good efficacy. No false positive and false negatives were detected when enriching digested DNA from human cell cultures and primary human cells. The protocol can probably be adapted to enriching sequences other than the hotspot sequence by altering the sequence in the capture oligonucleotide. We intend to apply this protocol in studies assessing effects of micronutrient status on meiotic recombination events in human sperm.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures




References
-
- Cheung VG, Sherman SL, Feingold E. Genetics. Genetic control of hotspots. Science. 2010;327:791–792. - PubMed
-
- Genome Browser, University of California Santa Cruz [07/06/2011];2009 http://genome.ucsc.edu/
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

