Oxidation-sensing regulator AbfR regulates oxidative stress responses, bacterial aggregation, and biofilm formation in Staphylococcus epidermidis
- PMID: 23271738
- PMCID: PMC3567629
- DOI: 10.1074/jbc.M112.426205
Oxidation-sensing regulator AbfR regulates oxidative stress responses, bacterial aggregation, and biofilm formation in Staphylococcus epidermidis
Abstract
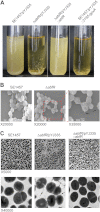
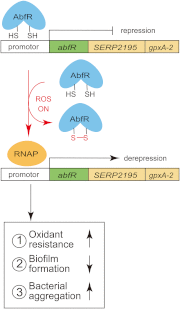
Staphylococcus epidermidis is a notorious human pathogen that is the major cause of infections related to implanted medical devices. Although redox regulation involving reactive oxygen species is now recognized as a critical component of bacterial signaling and regulation, the mechanism by which S. epidermidis senses and responds to oxidative stress remains largely unknown. Here, we report a new oxidation-sensing regulator, AbfR (aggregation and biofilm formation regulator) in S. epidermidis. An environment of oxidative stress mediated by H(2)O(2) or cumene hydroperoxide markedly up-regulates the expression of abfR gene. Similar to Pseudomonas aeruginosa OspR, AbfR is negatively autoregulated and dissociates from promoter DNA in the presence of oxidants. In vivo and in vitro analyses indicate that Cys-13 and Cys-116 are the key functional residues to form an intersubunit disulfide bond upon oxidation in AbfR. We further show that deletion of abfR leads to a significant induction in H(2)O(2) or cumene hydroperoxide resistance, enhanced bacterial aggregation, and reduced biofilm formation. These effects are mediated by derepression of SERP2195 and gpxA-2 that lie immediately downstream of the abfR gene in the same operon. Thus, oxidative stress likely acts as a signal to modulate S. epidermidis key virulence properties through AbfR.
Figures







References
-
- Uçkay I., Pittet D., Vaudaux P., Sax H., Lew D., Waldvogel F. (2009) Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41, 109–119 - PubMed
-
- Götz F. (2002) Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378 - PubMed
-
- Otto M. (2013) Staphylococcal infections. Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 64, 1–14 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

