Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis
- PMID: 23271803
- PMCID: PMC3545753
- DOI: 10.1073/pnas.1205750110
Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis
Abstract
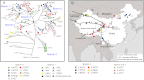
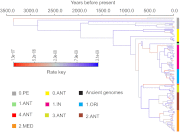
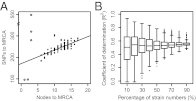
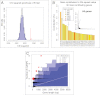
The genetic diversity of Yersinia pestis, the etiologic agent of plague, is extremely limited because of its recent origin coupled with a slow clock rate. Here we identified 2,326 SNPs from 133 genomes of Y. pestis strains that were isolated in China and elsewhere. These SNPs define the genealogy of Y. pestis since its most recent common ancestor. All but 28 of these SNPs represented mutations that happened only once within the genealogy, and they were distributed essentially at random among individual genes. Only seven genes contained a significant excess of nonsynonymous SNP, suggesting that the fixation of SNPs mainly arises via neutral processes, such as genetic drift, rather than Darwinian selection. However, the rate of fixation varies dramatically over the genealogy: the number of SNPs accumulated by different lineages was highly variable and the genealogy contains multiple polytomies, one of which resulted in four branches near the time of the Black Death. We suggest that demographic changes can affect the speed of evolution in epidemic pathogens even in the absence of natural selection, and hypothesize that neutral SNPs are fixed rapidly during intermittent epidemics and outbreaks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Gage KL, Kosoy MY. Natural history of plague: Perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. - PubMed
-
- Yersin A. La peste bubonique à Hong-Kong [Bubonic plague in Hong Kong] Ann Inst Pasteur (Paris) 1894;2:428–430.
-
- Benedictow OJ. The Black Death 1346–1353. Woodbridge: Boydell Press; 2004.
-
- Little LK. Plague and the End of Antiquity: The Pandemic of 541-750. New York: Cambridge Univ Press; 2007. p. xiv. 360.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

