Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections
- PMID: 23271804
- PMCID: PMC3549113
- DOI: 10.1073/pnas.1207025110
Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections
Abstract
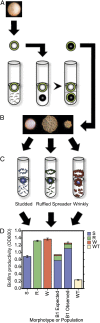
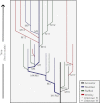
How diversity evolves and persists in biofilms is essential for understanding much of microbial life, including the uncertain dynamics of chronic infections. We developed a biofilm model enabling long-term selection for daily adherence to and dispersal from a plastic bead in a test tube. Focusing on a pathogen of the cystic fibrosis lung, Burkholderia cenocepacia, we sequenced clones and metagenomes to unravel the mutations and evolutionary forces responsible for adaptation and diversification of a single biofilm community during 1,050 generations of selection. The mutational patterns revealed recurrent evolution of biofilm specialists from generalist types and multiple adaptive alleles at relatively few loci. Fitness assays also demonstrated strong interference competition among contending mutants that preserved genetic diversity. Metagenomes from five other independently evolved biofilm lineages revealed extraordinary mutational parallelism that outlined common routes of adaptation, a subset of which was found, surprisingly, in a planktonic population. These mutations in turn were surprisingly well represented among mutations that evolved in cystic fibrosis isolates of both Burkholderia and Pseudomonas. These convergent pathways included altered metabolism of cyclic diguanosine monophosphate, polysaccharide production, tricarboxylic acid cycle enzymes, global transcription, and iron scavenging. Evolution in chronic infections therefore may be driven by mutations in relatively few pathways also favored during laboratory selection, creating hope that experimental evolution may illuminate the ecology and selective dynamics of chronic infections and improve treatment strategies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Phenotypic diversity and genotypic flexibility of Burkholderia cenocepacia during long-term chronic infection of cystic fibrosis lungs.Genome Res. 2017 Apr;27(4):650-662. doi: 10.1101/gr.213363.116. Epub 2017 Mar 21. Genome Res. 2017. PMID: 28325850 Free PMC article.
-
The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia.Mol Microbiol. 2011 Oct;82(2):327-41. doi: 10.1111/j.1365-2958.2011.07814.x. Epub 2011 Sep 7. Mol Microbiol. 2011. PMID: 21883527
-
Whole-Genome Sequencing of Three Clonal Clinical Isolates of B. cenocepacia from a Patient with Cystic Fibrosis.PLoS One. 2015 Nov 24;10(11):e0143472. doi: 10.1371/journal.pone.0143472. eCollection 2015. PLoS One. 2015. PMID: 26599356 Free PMC article.
-
Molecular approaches to pathogenesis study of Burkholderia cenocepacia, an important cystic fibrosis opportunistic bacterium.Appl Microbiol Biotechnol. 2011 Dec;92(5):887-95. doi: 10.1007/s00253-011-3616-5. Epub 2011 Oct 14. Appl Microbiol Biotechnol. 2011. PMID: 21997606 Review.
-
Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence.Clin Microbiol Infect. 2010 Jul;16(7):821-30. doi: 10.1111/j.1469-0691.2010.03237.x. Clin Microbiol Infect. 2010. PMID: 20880411 Review.
Cited by
-
The Rate and Molecular Spectrum of Spontaneous Mutations in the GC-Rich Multichromosome Genome of Burkholderia cenocepacia.Genetics. 2015 Jul;200(3):935-46. doi: 10.1534/genetics.115.176834. Epub 2015 May 12. Genetics. 2015. PMID: 25971664 Free PMC article.
-
Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post-transfer refinement.Elife. 2014 Nov 24;3:e04279. doi: 10.7554/eLife.04279. Elife. 2014. PMID: 25418043 Free PMC article.
-
Phenotypic diversity and genotypic flexibility of Burkholderia cenocepacia during long-term chronic infection of cystic fibrosis lungs.Genome Res. 2017 Apr;27(4):650-662. doi: 10.1101/gr.213363.116. Epub 2017 Mar 21. Genome Res. 2017. PMID: 28325850 Free PMC article.
-
Evolution of genotypic and phenotypic diversity in multispecies biofilms.NPJ Biofilms Microbiomes. 2025 Jul 1;11(1):118. doi: 10.1038/s41522-025-00755-1. NPJ Biofilms Microbiomes. 2025. PMID: 40595717 Free PMC article.
-
Identification of Cyclic-di-GMP-Modulating Protein Residues by Bidirectionally Evolving a Social Behavior in Pseudomonas fluorescens.mSystems. 2022 Oct 26;7(5):e0073722. doi: 10.1128/msystems.00737-22. Epub 2022 Oct 3. mSystems. 2022. PMID: 36190139 Free PMC article.
References
-
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. - PubMed
-
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. - PubMed
-
- O’Toole GA, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

