Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering
- PMID: 23271982
- PMCID: PMC3521655
- DOI: 10.1371/journal.pgen.1003134
Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering
Abstract
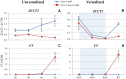
Most of the natural variation in wheat vernalization response is determined by allelic differences in the MADS-box transcription factor VERNALIZATION1 (VRN1). Extended exposures to low temperatures during the winter (vernalization) induce VRN1 expression and promote the transition of the apical meristem to the reproductive phase. In contrast to its Arabidopsis homolog (APETALA1), which is mainly expressed in the apical meristem, VRN1 is also expressed at high levels in the leaves, but its function in this tissue is not well understood. Using tetraploid wheat lines with truncation mutations in the two homoeologous copies of VRN1 (henceforth vrn1-null mutants), we demonstrate that a central role of VRN1 in the leaves is to maintain low transcript levels of the VRN2 flowering repressor after vernalization. Transcript levels of VRN2 were gradually down-regulated during vernalization in both mutant and wild-type genotypes, but were up-regulated after vernalization only in the vrn1-null mutants. The up-regulation of VRN2 delayed flowering by repressing the transcription of FT, a flowering-integrator gene that encodes a mobile protein that is transported from the leaves to the apical meristem to induce flowering. The role of VRN2 in the delayed flowering of the vrn1-null mutant was confirmed using double vrn1-vrn2-null mutants, which flowered two months earlier than the vrn1-null mutants. Both mutants produced normal flowers and seeds demonstrating that VRN1 is not essential for wheat flowering, which contradicts current flowering models. This result does not diminish the importance of VRN1 in the seasonal regulation of wheat flowering. The up-regulation of VRN1 during winter is required to maintain low transcript levels of VRN2, accelerate the induction of FT in the leaves, and regulate a timely flowering in the spring. Our results also demonstrate the existence of redundant wheat flowering genes that may provide new targets for engineering wheat varieties better adapted to changing environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

