The connectome mapper: an open-source processing pipeline to map connectomes with MRI
- PMID: 23272041
- PMCID: PMC3525592
- DOI: 10.1371/journal.pone.0048121
The connectome mapper: an open-source processing pipeline to map connectomes with MRI
Abstract
Researchers working in the field of global connectivity analysis using diffusion magnetic resonance imaging (MRI) can count on a wide selection of software packages for processing their data, with methods ranging from the reconstruction of the local intra-voxel axonal structure to the estimation of the trajectories of the underlying fibre tracts. However, each package is generally task-specific and uses its own conventions and file formats. In this article we present the Connectome Mapper, a software pipeline aimed at helping researchers through the tedious process of organising, processing and analysing diffusion MRI data to perform global brain connectivity analyses. Our pipeline is written in Python and is freely available as open-source at www.cmtk.org.
Conflict of interest statement
Figures


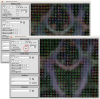
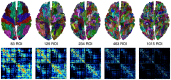
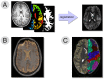
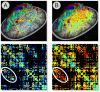

Similar articles
-
The development of brain connectivity browser by tractography of QBI.Annu Int Conf IEEE Eng Med Biol Soc. 2007;2007:2094-7. doi: 10.1109/IEMBS.2007.4352734. Annu Int Conf IEEE Eng Med Biol Soc. 2007. PMID: 18002400
-
An automated pipeline for constructing personalized virtual brains from multimodal neuroimaging data.Neuroimage. 2015 Aug 15;117:343-57. doi: 10.1016/j.neuroimage.2015.03.055. Epub 2015 Mar 31. Neuroimage. 2015. PMID: 25837600
-
The RUMBA software: tools for neuroimaging data analysis.Neuroinformatics. 2004;2(1):71-100. doi: 10.1385/NI:2:1:071. Neuroinformatics. 2004. PMID: 15067169
-
Automated processing pipeline for neonatal diffusion MRI in the developing Human Connectome Project.Neuroimage. 2019 Jan 15;185:750-763. doi: 10.1016/j.neuroimage.2018.05.064. Epub 2018 May 28. Neuroimage. 2019. PMID: 29852283 Free PMC article. Review.
-
Multiple-fiber reconstruction algorithms for diffusion MRI.Ann N Y Acad Sci. 2005 Dec;1064:113-33. doi: 10.1196/annals.1340.018. Ann N Y Acad Sci. 2005. PMID: 16394152 Review.
Cited by
-
Modeling time-varying brain networks with a self-tuning optimized Kalman filter.PLoS Comput Biol. 2020 Aug 17;16(8):e1007566. doi: 10.1371/journal.pcbi.1007566. eCollection 2020 Aug. PLoS Comput Biol. 2020. PMID: 32804971 Free PMC article.
-
Neurofilament light chain is increased in the parahippocampal cortex and associates with pathological hallmarks in Parkinson's disease dementia.Transl Neurodegener. 2023 Jan 20;12(1):3. doi: 10.1186/s40035-022-00328-8. Transl Neurodegener. 2023. PMID: 36658627 Free PMC article.
-
Deep brain stimulation of the internal capsule enhances human cognitive control and prefrontal cortex function.Nat Commun. 2019 Apr 4;10(1):1536. doi: 10.1038/s41467-019-09557-4. Nat Commun. 2019. PMID: 30948727 Free PMC article.
-
Altered white matter architecture in BDNF met carriers.PLoS One. 2013 Jul 31;8(7):e69290. doi: 10.1371/journal.pone.0069290. Print 2013. PLoS One. 2013. PMID: 23935975 Free PMC article.
-
Structural optimality and neurogenetic expression mediate functional dynamics in the human brain.Hum Brain Mapp. 2020 Jun 1;41(8):2229-2243. doi: 10.1002/hbm.24942. Epub 2020 Feb 6. Hum Brain Mapp. 2020. PMID: 32027077 Free PMC article.
References
-
- Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, et al. (1986) Mr imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 161: 401–407. - PubMed
-
- Basser PJ, Pierpaoli C (1996) Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. Journal of Magnetic Resonance, Series B 111: 209–219. - PubMed
-
- Basser PJ, Pajevic S, Pierpaoli C, Duda J, Aldroubi A (2000) In vivo fiber tractography using dt-mri data. Magn Reson Med 44: 625–632. - PubMed
-
- Mori S, van Zijl PC (2002) Fiber tracking: principles and strategies - a technical review. NMR Biomed 15: 468–480. - PubMed
-
- Hagmann P (2005) From diffusion MRI to brain connectomics. Ph.D. thesis, Ecole Polytechnique Fédérale de Lausanne.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

