Structural and functional analysis of multi-interface domains
- PMID: 23272073
- PMCID: PMC3522720
- DOI: 10.1371/journal.pone.0050821
Structural and functional analysis of multi-interface domains
Abstract
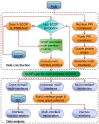
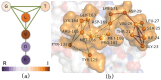
A multi-interface domain is a domain that can shape multiple and distinctive binding sites to contact with many other domains, forming a hub in domain-domain interaction networks. The functions played by the multiple interfaces are usually different, but there is no strict bijection between the functions and interfaces as some subsets of the interfaces play the same function. This work applies graph theory and algorithms to discover fingerprints for the multiple interfaces of a domain and to establish associations between the interfaces and functions, based on a huge set of multi-interface proteins from PDB. We found that about 40% of proteins have the multi-interface property, however the involved multi-interface domains account for only a tiny fraction (1.8%) of the total number of domains. The interfaces of these domains are distinguishable in terms of their fingerprints, indicating the functional specificity of the multiple interfaces in a domain. Furthermore, we observed that both cooperative and distinctive structural patterns, which will be useful for protein engineering, exist in the multiple interfaces of a domain.
Conflict of interest statement
Figures
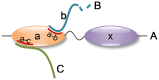

 and b represents
and b represents  .
.
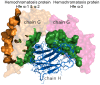
 -1 and
-1 and  2 domain (the left part of chain H) and Hemochromatosis protein Hfe
2 domain (the left part of chain H) and Hemochromatosis protein Hfe  -3 domain (the right part of chain H).
-3 domain (the right part of chain H).
Similar articles
-
The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation.Proc Natl Acad Sci U S A. 2012 Mar 6;109(10):3784-9. doi: 10.1073/pnas.1117768109. Epub 2012 Feb 21. Proc Natl Acad Sci U S A. 2012. PMID: 22355140 Free PMC article.
-
PIBASE: a comprehensive database of structurally defined protein interfaces.Bioinformatics. 2005 May 1;21(9):1901-7. doi: 10.1093/bioinformatics/bti277. Epub 2005 Jan 18. Bioinformatics. 2005. PMID: 15657096
-
Oligomeric protein structure networks: insights into protein-protein interactions.BMC Bioinformatics. 2005 Dec 10;6:296. doi: 10.1186/1471-2105-6-296. BMC Bioinformatics. 2005. PMID: 16336694 Free PMC article.
-
Characterization and prediction of protein interfaces to infer protein-protein interaction networks.Curr Pharm Biotechnol. 2008 Apr;9(2):67-76. doi: 10.2174/138920108783955191. Curr Pharm Biotechnol. 2008. PMID: 18393863 Review.
-
Protein interactions in 3D: from interface evolution to drug discovery.J Struct Biol. 2012 Sep;179(3):347-58. doi: 10.1016/j.jsb.2012.04.009. Epub 2012 May 1. J Struct Biol. 2012. PMID: 22595401 Review.
References
-
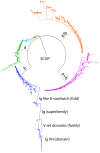
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540. - PubMed
-
- Kim PM, Lu LJ, Xia Y, Gerstein MB (2006) Relating three-dimensional structures to protein networks provides evolutionary insights. Science 314: 1938–1941. - PubMed
-
- Dasgupta B, Nakamura H, Kinjo AR (2011) Distinct Roles of Overlapping and Non-overlapping Regions of Hub Protein Interfaces in Recognition of Multiple Partners. J Mol Biol 411: 713–727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

